
Towards Confidentiality-strengthened Personalized Genomic Medicine
Embedding Homomorphic Cryptography
Kalpana Singh
1
, Renaud Sirdey
1
, Franc¸ois Artiguenave
2
, David Cohen
3
and Sergiu Carpov
1
1
CEA, LIST, Point Courrier 172, 91191 Gif-sur-Yvette Cedex, France
2
TRAASER, 4 rue Pierre Fontaine, 91028 Evry, France
3
CEA, CNG, 91057 Evry Cedex, France
{kalpana.singh, renaud.sirdey, sergiu.carpov}@cea.fr, francois.artiguenave@traaser.com, cohen@cng.fr
Keywords:
Genome Sequencing, Personalized Medicine, Privacy, Homomorphic Encryption.
Abstract:
In this paper, we analyze and propose a solution for the challenges that come with personalized genomic and,
most importantly, of performing queries on sequenced dataset sitting on a cloud server. This work provides
scenarios for its application in personalized genomic medicine, and tests homomorphic encryption technique
to assist in improving the strength of their privacy at non-prohibitive performance cost. By experimental
testing using HElib, we make a first step towards performing practical computation over the relevant portions
of the genomic dataset of an individual for a first round of practical diagnosis rules.
1 INTRODUCTION
Physicians and researchers think that understanding
how genes influence disease will require genetic and
health datasets to be collected from millions of peo-
ple. Such a massive task will probably require har-
nessing the processing power of networked cloud
computers, but online security breaches in the past
few years illustrate the dangers of entrusting huge,
sensitive datasets to the cloud. Genomic dataset anal-
ysis is increasingly incorporated in a variety of do-
mains, including personalized medicine, biomedical
research, direct-to-consumer services, and forensics.
Genome sequencing technology has advanced at
a rapid pace and it is now possible to generate
highly-detailed genotypes inexpensively. As a re-
sult, genome sequencing may soon become a routine
tool for clinical diagnosis and therapy selection. In
the (near) future, personalized medicine will result in
“right drug at the right time” according to their pa-
tients’ genome dataset.
The implementation of genomic-based medicine
is the challenge of transmitting clinically useful
information to health-care practitioners. In this study,
we choose personalized medicine case scenario as
a model setting for an implementation of privacy-
preserving genetic testing and results reporting.
Genomic Dataset Privacy Issues. The whole
genome sequencing was initiated at the U.S. National
Institutes of Health (NIH) in 1990 and the first full
sequence was released years later at a total cost of
$3 billion. Yet, sequencing technology has evolved
and costs have plummeted, such that the price for a
whole genome sequence is $5K as of July 2014 and
can be completed in two to three days. The “$1K
genome in 1 day” will soon be a reality (Naveed
et al., 2015). As the cost of sequencing the human
genome drops, more and more genomic dataset will
become available for research and study. At the
same time, researchers are developing new methods
for analyzing genomic dataset across populations
to look for patterns and find correlations. Such
research may help identify genetic risk factors for
diseases, suggest treatments, or find cures. To make
this dataset available for scientific study, patients
expose themselves to risks from invasion of privacy
(Ayday et al., 2013a). Decreases in sequencing
costs have coincided with an escalation in genomics
as a research discipline with explicit application
possibilities.
Personalized Medicine. Personalized medicine
promises to revolutionize healthcare through treat-
ments tailored to an individual’s genomic makeup and
genome-based disease risk tests that can enable early
diagnosis of serious diseases such as diagnosis of sus-
pected mendelian conditions and for targeting cancer
treatments. The current rise of personalized medicine
is based on increasing affordability and availability of
Singh, K., Sirdey, R., Artiguenave, F., Cohen, D. and Carpov, S.
Towards Confidentiality-strengthened Personalized Genomic Medicine Embedding Homomorphic Cryptography.
DOI: 10.5220/0006148303250333
In Proceedings of the 3rd International Conference on Information Systems Security and Privacy (ICISSP 2017), pages 325-333
ISBN: 978-989-758-209-7
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
325

individual genome sequencing.
As mentioned in Subsection 1, the cost to se-
quence an entire human genome continues to fall, the
potential exists for rapid advances in wellness and
health care resulting from this new technology. Es-
sential to achieving those advances is the need to out-
source, compare, and aggregates the genome dataset.
However, as the ease with which the acquisition and
outsourcing of genome sequencing information in-
creases, so we will have questions and concerns about
privacy, security, and efficiency.
1.1 Contributions of this Paper
In this paper, we propose an architecture and its
application in personalized medicine case scenario.
We test homomorphic encryption techniques to as-
sist in improving the strength of their privacy at
non-prohibitive performance cost. We experimen-
tally analyse our personalized medicine case sce-
nario architecture using the HElib library, and HElib
achieves near practical computation cost. We show
that the proposed solution have used real genomic
rules, which have been generated by the geneticists
in our team (authors 2 and 3). Our main contribu-
tions are (i) to keep genomic datasets secure while
still enabling cloud-based analyses needed to make
meaningful diagnosis, (ii) provide acceptable level
of privacy requirements in each step of handling of
genomic datasets, collecting, analyzing, storing or
sharing the genetic informations and (iii) provide a
characterization of various threat models that are ad-
dressed at each step.
The paper is organized as follows. In Section 2,
we review the current literature relative to the chal-
lenges mentioned in Section 1. Section 3 provides an
architecture, including descriptions of various main
components. Section 4 gives a detailed description of
homomorphic encryption method used in our method-
ology. Section 5 presents an insider threat model for
our architecture. Section 6 demonstrates the exper-
imental results, and indicates homomorphic encryp-
tion overhead is not prohibitive for this application.
Section 7 summarizes and presents conclusions.
2 THE CURRENT SOLUTIONS
Privacy issues caused by forensic, medical and other
uses of genomic dataset have been studied in the past
few years (Jiang et al., 2014), and (Naveed et al.,
2015). Homomorphic encryption technique is quickly
becoming more relevant due to its great potential for
privacy computation on encrypted genomic datasets.
This technique has a number of other advantages, al-
lowing for more flexible case scenarios, and requiring
less interaction, thereby reducing the communication
complexity. The cryptographic overhead consists of
the time to perform operations for each gate of the
circuit as well as other maintenance operations. Un-
fortunately, it is hard to characterize simply the cryp-
tographic overhead of fully homomorphic encryption
(FHE) because there are a lot of parameters that affect
its performance, such as the multiplicative depth, the
security parameter, the plaintext size, the exact FHE
scheme used, the performance of various operations
in the finite fields used. Lepoint and Naehrig (Le-
point and Naehrig, 2014) and Halevi (Halevi, 2013)
provide performance measurements for various set-
tings of these parameters. A number of key optimiza-
tions and batch techniques (Halevi and Shoup, 2014),
(Zhou and Wornell, 2014) have been introduced to re-
duce overall computation complexity and increase ef-
ficiency of these homomorphic based schemes. Re-
cently, homomorphic encryption techniques (Ayday
et al., 2014), (Ayday et al., 2013b), (Lauter et al.,
2015) have been used to encrypt genomic datasets in
such a way that storage can be outsourced to an un-
trusted cloud, and the datasets can be computed on
in a meaningful way in encrypted form, without re-
quiring access to decryption keys. These protocols
have some drawbacks such as being computationally
intensive, leaking more than necessary and being un-
scalable; mainly due to the very large size of genomic
datasets. However, a number of optimization tech-
niques (Halevi and Shoup, 2014), (Zhou and Wornell,
2014) have been presented to overcome the limita-
tions of using homomorphic based solutions. Build-
ing practical systems that compute on encrypted ge-
nomic datasets are a challenging task. One reason
is that homomorphic encryption method remains too
slow for running arbitrary functions or for enabling
the complex systems we have today. Another rea-
son is that many systems take advantage of fast search
data structures (such as database indexes), and a prac-
tical system must preserve this performance over en-
crypted dataset.
We present a cryptographic solution for genomic
datasets storage and outsourcing, and maintaining
patient privacy. All encrypted genomic datasets
are stored in an untrusted cloud server. To allow
meaningful computation on the encrypted genomic
datasets, we use HElib (Halevi, 2013), (Halevi and
Shoup, 2014). Specially, we take basic genomic algo-
rithms which are commonly used in genetic associa-
tion studies and show how they can be made to work
on encrypted genotype and phenotype datasets. We
also tackled the insider attack situation in an untrusted
ICISSP 2017 - 3rd International Conference on Information Systems Security and Privacy
326
cloud server where the attacker is assumed to have ac-
cess to the content of the disk as well as the CPU and
the memory.
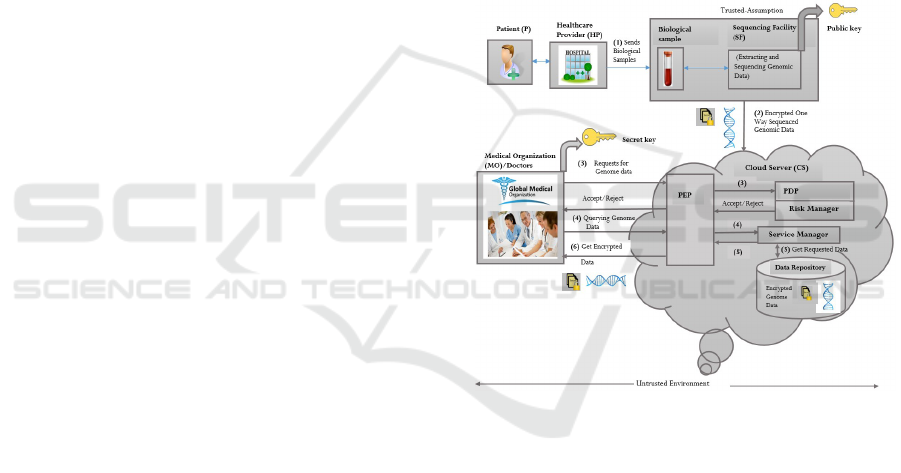
3 OUR ARCHITECTURE
The system architecture is designed for enhanced pri-
vacy protection of encrypted genomic datasets down-
loaded to a medical organization/doctor from the
cloud server, such as in the case of personalized
medicine. The proposed system (Figure 1) involves
five components: (i) the patients; (ii) Healthcare
Providers (HPs) have patients’ biological samples;
(iii) a Sequencing Facility (SF) responsible for se-
quencing, management of cryptographic keys, and
encryption of patients’ sequenced genomic datasets;
(iv) a Cloud Server (CS) where the encrypted form
of “sequenced genomic datasets” (EGDS) are stored,
and all computations and communications held on;
and (v) Medical Organizations (MOs), or doctors,
wishing to perform genetic tests on the patients’ ge-
nomic datasets. Our full architecture includes sev-
eral patients, several HPs, and several MOs/doctors,
all accessing the one server. The CS has a compo-
nent which we refer to as Service Manager (SM),
which is responsible for dataset storage in the CS,
performs computation on requested query to get the
result, and sends query result to the MO/doctor. To
simplify the explanations, in the system model here
we present only one patient, one HP, one MO/doctor,
as shown in Figure 1. In our architecture, MO/doctor
is a part of the HP. The MO/doctor has the secret key
to decrypt EGDS and get the sequenced EGDS for
further analysis and research. Patient can also have
secret key to get the EGDS from the CS. To secure
the communication, the participants (patient, a HP,
and the MO/doctor) are authenticated, and the con-
nection is protected using authorization process and
risk management policies. We deploy access con-
trol policies at the CS. The CS performs access con-
trol by making decision requests and enforcing access
control decisions using Policy Enforcement Points
(PEPs) (Q.Yaseen et al., 2013). The system entities
that evaluate the applicable policies and make an ac-
cess control decision are referred to as Policy Deci-
sion Points (PDPs) (Q.Yaseen et al., 2013).
In the following sections, we describe in detail
the components of our architecture. We can classify
five components of our architecture into three mod-
ules. These three modules are data contributor, cloud
server, and data requester. Data contributor has three
components; a patient, a HP, and a SF. Data requester
is a MO or doctor. Our architecture is depicted in Fig-
ure 1, which shows an interaction between a patient,
a HP, SF, a MO/doctor, and a CS.
3.1 Data Contributor Module
The data contributor module has three components: a
patient, a HP, and a SF.
Patient. Each patient has history of medical records
and biological samples to the HP, which is containing
sensitive data such as blood samples. Patient has the
secret key to see the content of his EGDS, which is
stored on the CS.
Healthcare Provider (HP). The HP is responsible for
good medical treatment of each patient. The HP sends
biological samples such as blood sample to the SF, in
order to sequence genome datasets for better medical
treatment.
Figure 1: The Proposed Architecture - A Personalized
Medicine Case Scenario.
Sequencing Facility (SF). The main component on
the data contributor is the SF. Now, the SF has bio-
logical samples, which is received from the HP for
sequencing analysis. We note that the SF currently
would have access to unprotected raw genetic vari-
ants and therefore must be a trusted entity. The SF
is responsible for genome sequencing received from
the HP, encrypting genome datasets using strong sym-
metric data encryption, and sending encrypted form
of sequenced genomic datasets (SGDS) to the CS for
dataset storage. In addition, we are assuming a model
where the encrypted SGDS are stored in a central-
ized untrusted CS rather than at the HP, which maxi-
mizes efficiency and security. To maintain privacy of
SGDS on the CS with no storage overhead, transci-
phering techniques (Canteaut et al., 2016) have to be
used to translate these datasets into the homomorphic
Towards Confidentiality-strengthened Personalized Genomic Medicine Embedding Homomorphic Cryptography
327

domain, where they can be processed with respect to
the public key of a given MO/doctor (that way, FHE
overhead is paid only transiently in the server memory
during the homomorphic calculation). In this scheme,
each individual ciphertext element is conceptually a
vector of encrypted plaintext integrals. This construc-
tion gave rise to a SIMD style operations that could
particularly be effective with problems that benefit
from some level of parallel computation.
3.2 Cloud Server Module
Cloud computing provides massive computation
power and storage capacity which enables a user to
deploy applications without infrastructure investment
(Zhang et al., 2013). We do not deal with the CS’s in-
ternal working here (See Figure 1). The service man-
ager (SM) is a function of the CS which handles each
send/receive process on a CS. The SM accepts EGDS
from the SF and stores in the data repository. The SM
privately processes queries, gives ‘private’ feedback
to MO or doctor, and maintains the credential revo-
cation. At the CS, we also use access control mech-
anisms to provide additional layer of security for in-
sider threat.
Risk Assessment. Insiders use many approaches and
factors to launch attacks. Two of the most risky fac-
tors that can be used are insiders’ knowledgebase and
dependencies among datasets. Modern access control
mechanisms use the request-response paradigm. This
model consists of the Policy Decision Point (PDP)
(Q.Yaseen et al., 2013), which is responsible of issu-
ing accessing decisions, and the Policy Enforcement
Point (PEP) (Q.Yaseen et al., 2013), which is respon-
sible of enforcing these decisions. We adopted these
control policies in our architecture. Adding policies
will increase cost in our architecture. So, we need a
solution to this problem should take into account the
trade off between preventing insider threat and perfor-
mance.
The SM is responsible for each send and receive
process on the CS by means of the traditional request-
response paradigm, in which policies are established
and decisions are made on the basis of these poli-
cies. In recent papers encouraging the development
of a standard approach to risk management in cloud
services and grids. The European Grid Infrastructure
(EGI) design as appearing at (https://www.egi.eu/),
now establishes a standard for risk evaluation and mit-
igation. Figure 4 of the paper (Nogoorani and Jalili,
2016) illustrates an architecture providing risk pre-
vention by means of a separation of policy decision
from enforcement, along with a thorough evaluation
of trust and risk. We do not require the extensive risk
assessment of the EGI, but do follow its recommenda-
tions by separating PEP from PDP and by introducing
a risk manager at the PDP side in our architecture for
preventing insider threat in PEP-side caching model
while keeping a low overhead on PEP and PDP per-
formance.
3.3 Data Requester Module
The component of the data requester includes decryp-
tion process of BGV method (see in Section 4) to get
requested data. The MO/doctor requests data query
to access EGDS from the CS, the CS sends requested
data in encrypted form. The MO/doctor has secret key
to decrypt data using “Decryption steps” from BGV
method and get the requested data. Data queries of
medical significance only exploit a small portion of
genome data (typically a few tens of positions com-
bined in a boolean expression). Hence, as long as the
storage overhead issue is solved using transciphering
techniques the practical processing of phenotype de-
termination queries in an encrypted domain appears
achievable (or almost so) using present day homomor-
phic encryption techniques.
4 THE BGV SCHEME
The Brakerski-Gentry-Vaikuntanathan (BGV)
scheme (Brakerski et al., 2012) stands today as one
of the most efficient somewhat FHE scheme. The
implementation of this scheme has been discussed
in the literature (Brakerski et al., 2012), which is
focused on the evaluation of AES (Gentry et al.,
2010). In our setup, it involves encrypting genomic
datasets on a SF, then uploading EGDS to the CS.
Computations on EGDS are performed in the CS and
an encrypted result is then sent back to a MO/doctor
as requested data from the CS. The MO/doctor
decrypts an answer using secret key. If attackers were
to intercept EGDS at any point along the way, the
underlying data would remain safe. We use HElib
software library for our experimental analysis and
results (see in Subsection 6.2), which implements
the BGV scheme, along with many optimizations to
make homomorphic evaluation runs faster.
Brakerski-Gentry-Vaikuntanathan (BGV)
Scheme. This section provides a basic descrip-
tion of BGV scheme.
Notation Descriptions. Let us denote by [.]
q
the re-
duction modulo q into the interval (−q2;q2] ∩
Z of the integer or integer polynomial (coefficient-
wise). For a security parameter λ, we choose an
ICISSP 2017 - 3rd International Conference on Information Systems Security and Privacy
328

integer m = m(λ) that defines the m-th cyclotomic
polynomial φ
m
(x). For a polynomial ring R =
Z[x](φ
m
(x)), set the plaintext space to R
t
:= RtR)
for some fixed t ≥ 2 and the ciphertext space to R
q
:=
RqR for an integer q = q(λ). Let X = X(λ) denotes
a noise distribution over the ring R. We use the stan-
dard notation a ← D to denote that a is chosen from
the distribution D. Now, we recall the BGV scheme
(Brakerski et al., 2012). Gentry, Halevi and Smart
(Gentry et al., 2010) constructed an efficient BGV
scheme. The security of this scheme is based on the
(decisional) Ring Learning With Errors (RLWE) as-
sumption. The assumption is that it is infeasible to
distinguish the following two distributions. The first
distribution consists of pairs (a
i
,u
i
), where a
i
, u
i
←−
R
q
uniformly at random. The second distribution con-
sists of pairs of the form (a
i
,b
i
) = (a
i
,a
i
s + e
i
) where
a
i
←− R
q
drawn uniformly and s, e
i
←− X. Note
that we can generate RLWE samples as (a
i
,a
i
s +te
i
)
where t and q are relatively prime. To improve effi-
ciency for HE, they use very sparse secret keys s with
coefficients sampled from {−1,0, 1}. Here is the SHE
scheme of (Brakerski et al., 2012):
ParamsGen: Given the security parameter λ,
choose an odd integer m, a chain of moduli q
0
<
q
1
< ........ < q
L−1
= q, a plaintext modulus t with
1 < t < q
0
, and discrete Gaussian distribution X
err
.
Output (m,{q
i
},t,X
err
).
KeyGen: On the input parameters, choose a ran-
dom s from {0, ±1}
φ(m)
and generate an RLWE in-
stance (a,b) = (a,[as + te]
q
) for e ← X
err
. For an in-
teger P, we define the key switching matrix W =
b
s
a
s
where b
s
= [a
s
.s + te
s
+ Ps
2
]
P
q
L−2
for a
s
← R
q
uniformly at random and e
s
← X
err
.
We set the key pair:(pk,sk, evk) = ((a,b), s,W ). Then
we define the SwitchKey(c,evk) for the extended ci-
phertext c = (d
0
,d
1
,d
2
) at level l as follows: set
c
0
=
c
0
0
c
0
1
=
"
Pd
0
[b
s
]
P
ql
Pd
1
[a
s
]
P
ql
1
d
2
#
P
ql
, (1)
and then take an element c
00
∈ R
ql
such that c
00
≡
c
0
(modt) and c
00
is the closet to P.c
0
modulo t.
Encryption: To encrypt m ∈ R
t
, choose a small
polynomial v and two Gaussian polynomials e
0
, e
1
over R
q
. Then compute the ciphertext given by
Enc(m, pk) = (c
0
,c
1
) = (m,0) + (bv +te
0
,av + te
1
) ∈ R
2
q
(2)
Decryption: Given a ciphertext ct = (c
0
,c
1
) at
level l, output Dec(ct, sk) = [c
0
− s.c
1
]
ql
mod t where
the polynomial [c
0
− s.c
1
]
ql
is called the noise in the
ciphertext ct.
Homomorphic Evaluation: Given two cipher-
texts ct = (c
0
,c
1
) and ct
0
= (c
0
0
,c
0
1
) at level l, the
homomorphic addition is computed by ct
add
= ([c
0
+
c
0
0
]
ql
,[c
1
+ c
0
1
]
ql
). The homomorphic multiplication is
computed by ct
mult
= SwitchKey(c
0
∗ c
1
,evk) where
c
0
∗c
1
= ([c
0
c
0
0
]
ql
,[c
0
c
0
1
+c
1
c
0
0
]
ql
,[c
1
c
0
1
]
ql
) and the key
switching function SwitchKey is used to reduce the
size of ciphertexts to two ring elements. We also ap-
ply modulus switching from q
i
to q
i−1
in order to
reduce the noise. If we reach the smallest modulus
q
0
, we can no longer compute on ciphertexts. BGV
scheme adapted the Smart and Vercauteren scheme
(Smart and Vercauteren, 2014) for optimization.
5 INSIDER THREAT MODEL
This section addresses that we prevent the CS from
launching the insider attacks. In our architecture, the
SM manages and stores EGDS from the SF. These
datasets may have dependencies that can be deter-
mined by an insider. In addition insiders can retrieve
general knowledge about the types of datasets being
stored. Use of such dependencies among datasets and
the knowledgebase of insiders (Nogoorani and Jalili,
2016), (Q.Yaseen et al., 2013) may enable the insider
to infer sensitive information. We choose the follow-
ing criteria for the establishment of protection against
insider attacks:
1. To prevent the SM from reading plaintext ge-
nomic datasets. As presented in Subsection 3.2, the
SF employs computationally strong homomorphic en-
cryption that makes the possibility of an exposure
negligible even if insiders get access to the encrypted
datasets. To prevent any kind of information leakage,
we use the whole datasets are encrypted. The SM per-
forms all computation on the EGDS. Since the origi-
nal dataset always remains encrypted on the CS, there
is no direct way in which the insider can access this
data unless he is able to break the encryption method.
With the privacy model and the analysis of attacks,
we find that our EGDS is strongly secure from the in-
siders.
2. We did not present the query processing steps
between the CS and the MO/doctor in detail in this pa-
per. However, based on the formal model for security
of BGV scheme provides privacy of the MO/doctor
as long as the underlying homomorphic encryption
scheme is secure.
3. Prevention of abuse of PEP-side caching in
Sub-subsection 3.2, we describe the risks involved be-
cause of the standard caching methods used in the CS.
This can be resolved by a separation of the PDP from
the PEP as described in (Q.Yaseen et al., 2013), along
Towards Confidentiality-strengthened Personalized Genomic Medicine Embedding Homomorphic Cryptography
329

with introduction of an insider threat detection unit at
the PDP side is used in our scheme to prevent PEP-
side caching; we have implemented this in our archi-
tecture. The authors of (Nogoorani and Jalili, 2016)
also use this separation to prevent this insider attack
(see their Figure 4).
6 EXPERIMENTAL RESULTS
This section presents the computation cost analysis
of genomic datasets encryption, computation and
decryption time. Our objective is to prove that
the computation cost of homomorphic encryption
scheme is close to be practical. We explain how
to set the parameters for homomorphic evaluations
and present our experimental results. We use HElib
software library that implements BGV scheme.
HELib is written in C++ and based on the arithmetic
library NTL (http://www.shoup.net/ntl/) over GMP
(https://gmplib.org/).
HElib Library. Due to use of BGV system we
can evaluate many such instances in parallel using
batching. We further use additional optimization
including a systematic use tree-structured multiplica-
tions to achieve very low multiplicative depth on the
specific kind of algorithms involved in our study.
6.1 Datasets and ABO Rules
We utilize the public available dataset Blood Group
Antigen Gene Mutation Database (BGMUT), which
is an online repository of allelic variations in genes
that determine the antigens of various human blood
group systems. Currently, the database documents
sequence variations of a total of 1251 alleles of all
40 gene that together are known to affect antigens
of 30 human blood group systems. BGMUT is a
part of the dbRBC resource of the National Center
for Biotechnology Information, and is available at
(https://www.ncbi.nlm.nih.gov/projects/gv/mhc/xslcg
i.cgi?cmd=bgmut/home).
We develop two ABO rules using BGMUT
datasets. We run our HElib-based prototype on these
datasets and evaluate our ABO rules to see the pres-
ence of ABO blood type for each patient. We have
two ABO rules labeled as ABO-1, and ABO-2. The
size of ABO-1 is 2.79 KB which is very small in com-
parison with ABO-2 size, which is 45.4 KB (approx.
23 pages in .doc extension, 11 pt “times new roman”
size of content). A total of 2504 patients are included
in this study. The size of each patient dataset is 17.7
KB, in total we have 44.3 MB datasets for the exper-
imental analysis. Each dataset is defined in table for-
mat, which has six columns. The sample of each pa-
tient’s dataset is defined in Table 1.
Table 1: Sample of Datasets.
Chromo-
some
Position RC AA Values-1 Values-2
9 136125819 C T 0 0
9 136126129 A C 0 1
Note: Where, RC - Reference Chromosome, AA - Alternative Allele.
In first row of Table 1, the “9” indicates chro-
mosome, an integer number “136125819” that corre-
sponds to the position at the reference chromosome,
“C” which is the nucleotide at the reference chromo-
some, and “T” which indicates the nucleotides of the
alternative allele. In the columns 5 and 6, both values
are “0”, which means that the both alleles of an in-
dividual have no change in the nucleotide compared
to reference chromosome. Similarly, all the rows are
defined. In second row, values are “0” and “1”, which
mean that one of the alleles has a polymorphism at
that specific chromosome position. If we have both
values are “1” means that both alleles have that spe-
cific polymorphism.
6.2 Experiment Setup and Results
This section presents the experimental results of ABO
rules using HElib. Our experiments using HElib are
performed on an Ubuntu 14.04 virtual box which is
running on windows 7 with Intel (R) Core (TM) i5-
5300U CPU, 2.30 GHz processor, and RAM 16 GB.
We present performance costs in the terms of time and
size. We carried out 20 runs for each experiment to
obtain the average performance metric values.
6.2.1 HElib Parameter Settings
We present the HElib parameter setting for secure ge-
nomic datasets analysis in Table 3. The HElib context
is mainly defined by the plaintext base (p), the secu-
rity parameter (λ) and the circuit depth (L). All the
parameters provide 128-bit security level. In particu-
lar, L can be considered as the number of ciphertext
moduli in the BGV scheme. We choose the following
parameter values: p = 2 and λ = 128. The values of
L are presented for evaluation of both rules in Table
3. The security of BGV relies on the hardness of the
RLWE assumption (Brakerski et al., 2012).
6.2.2 Experimental Results
We measure the running time of key generation, en-
cryption, evaluation, and decryption to evaluate the
ICISSP 2017 - 3rd International Conference on Information Systems Security and Privacy
330

overall performance as well as the impact of the HE-
lib in our considered genome datasets. This segment
presents the experimental results of both rules, which
are shown in Tables 3, and 4 respectively. In our
datasets, each of genome sequence is expressed in a
binary representation.
ABO-1 Rule: We implement and analyze the com-
putation cost of running ABO-1 rule on genomic
datasets. Our ABO-1 rule is presented in Table 2. In
ABO-1, first column indicates the blood group type
and the second column indicates the conditions that
are required to have that specific blood group type.
We define 15 rules (O1O1, A2O1, A2A2, B1O1,
B1B1, A2B1, A1O1, O1O3, A1A1, A1O3, O3O3,
A1B1, B1O3, A1A2, A2O3), which are presented in
Table 2. For example, in order to have the blood group
type O1O1, the individual should not have an poly-
morphism at chromosome 9 at position 136132908;
indicated by the factor (9:136132908;T;TC;0|0).
Table 2: Our ABO-1 Rule.
Phenotype Rule
O1O1 (9:136132908;T;TC;0|0)
A2O1 (9:136132908;T;TC;0|1) & (9:136131651;G;A;0|1)
A2A2 (9:136132908;T;TC;1|1) & (9:136131651;G;A;1|1)
B1O1 (9:136132908;T;TC;0|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|1)
B1B1 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;1|1)
A2B1 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|1) &
(9:136131461;G;A;0|1)
A1O1 (9:136132908;T;TC;0|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|0) & (9:136131316;C;T;0|0)
O1O3 (9:136132908;T;TC;0|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|0) & (9:136131316;C;T;0|1)
A1A1 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|0) & (9:136131316;C;T;0|0)
A1O3 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|0) & (9:136131316;C;T;0|1)
O3O3 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|0) & (9:136131316;C;T;1|1)
A1B1 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|1) & (9:136131316;C;T;0|0)
B1O3 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|0) &
(9:136131461;G;A;0|1) & (9:136131316;C;T;0|1)
A1A2 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|1) &
(9:136131461;G;A;0|0) & (9:136131316;C;T;0|0)
A2O3 (9:136132908;T;TC;1|1) & (9:136131651;G;A;0|1) &
(9:136131461;G;A;0|0) & (9:136131316;C;T;0|1)
In Table 2, “9” indicates chromosome, fol-
lowed by a separator “:” and an integer number
“136131316” that corresponds to the position at the
reference chromosome. The “;” is separator followed
by “T” which is the nucleotide at the reference
chromosome, followed by a separator “;” and then
the value “T” which indicates the nucleotides of the
alternative allele. The difference between the refer-
ence chromosome and the sample chromosome: the
sample/individual contains an additional nucleotide
“C” with respect to the reference. In case it is written
“;G;A;” it means that in the individual the nucleotide
G has been changed into the nucleotide A with respect
to the reference chromosome. Another separator “;”
followed by the genotype (GT) annotation. 0|0 means
that both alleles of an individual have no change in
the nucleotide compared to reference chromosome.
0|1 means that one of the alleles has a polymorphism
at that specific chromosome position. 1|1 means that
both alleles have that specific polymorphism and thus
the individual is homozygote for that polymorphism.
ABO-2 Rule: This segment describes the ABO-2
rule. This rule contains many sections to determine
haplotype and phenotype (A, B, AB and O). One
section contains simple rules that can determine
phenotype but this prediction is not 100% accurate
as it leaves out certain very rare alleles. The other
section contains more complicated rules that take all
(known) alleles into account and this should be more
accurate.
The size of ABO-2 rule is presented in Subsec-
tion 6.1, as the size of ABO-2 is quite large, which
is not possible to present in this paper. We develop
ABO-2 rule for determining these blood groups for
each patient such as: O haplotype, B haplotype, T
haplotype. Additionally, we develop extended rules
to decrease (false negative) FN and (false positive) FP
for these rules O haplotype, B haplotype, and T haplo-
type. We introduce simple rules for determining phe-
notypes such as O phenotype, T phenotype (T pheno-
type w/ TT genotype, T phenotype w/TO genotype),
B phenotype (B phenotype w/BB genotype, B pheno-
type w/BO genotype, TB phenotype). At the last, we
develop some extended rules for determining pheno-
type rules with genotype including B phenotype rules
for genotype, T phenotype rules for genotype, and TB
phenotype w, corresponding rules for genotype.
We implement these ABO-1, and ABO-2 rules us-
ing HElib to identify the presence of ABO blood types
in patient’s blood for further analysis and better treat-
ment. The results are shown in Tables 3, and 4 respec-
tively. Table 3 describes the parameters which have
been used in Table 4 for our experimental analysis.
We use default value of plaintext base (p) which is
2. The value of circuit depth (L) depends on the num-
ber of multiplications in ABO-1, and ABO-2 rules.
We use tree-structured multiplications to achieve low
multiplicative depth in evaluation of ABO-1, and
ABO-2 rules.
Tables 3, and 4 present one patient dataset analy-
sis. We can run these experiments using paralleliza-
Towards Confidentiality-strengthened Personalized Genomic Medicine Embedding Homomorphic Cryptography
331
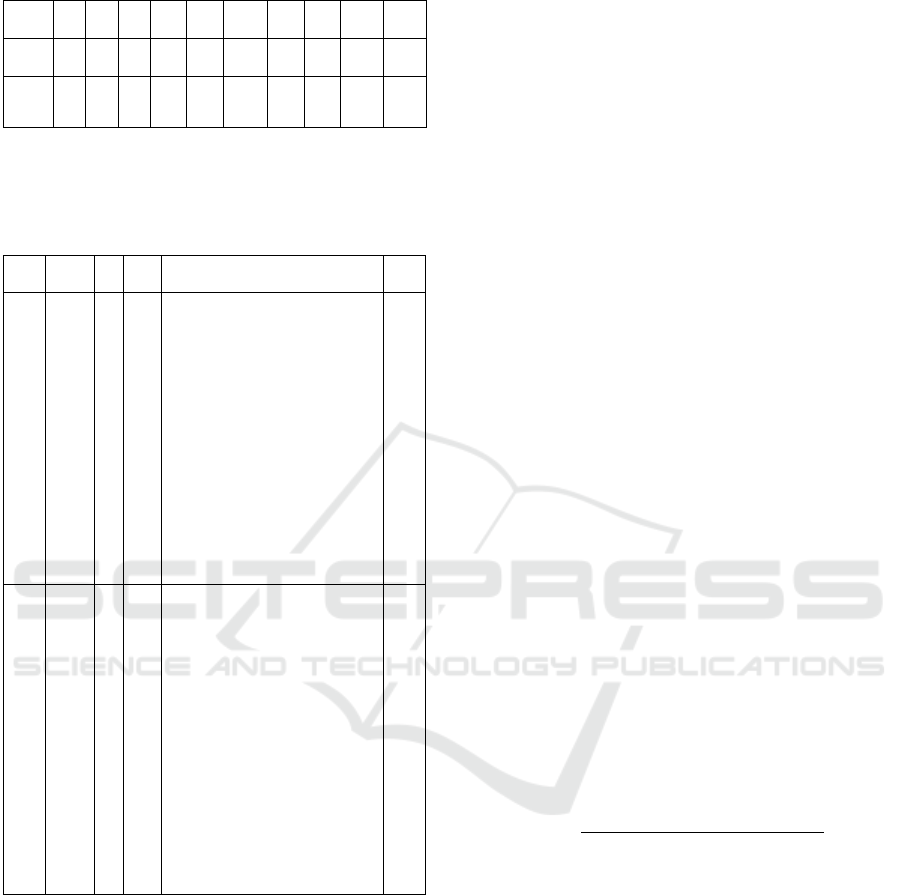
Table 3: HElib Parameters for ABO rules Execution.
ABO
Rules
p L BL λ OT CT PK SK NoM NoA
ABO-
1
2 5 4 128 17.7
KB
851.6
KB
29.0
MB
29.2
MB
7 7
ABO-
2
2 15 14 128 17.7
KB
22300
KB
93.4
MB
94.6
MB
20
60
29
53
17
Note: Where, L - Circuit Depth, BL- Base Level, λ- Security in Bits, OT- Original Text in
KiloBytes, CT- Cipher Text in KiloBytes, PK- Public Key in MegaBytes, SK-Secret Key
in MegaBytes, NoM - No. of Multiplications, NoA - No. of Additions.
Table 4: Sequential Time cost in Seconds for ABO rules
Execution.
ABO
Rules
KG E Eval D TE
ABO-
1
2.732 0 3
Decrypted Value for rule O1O1 is: 0
Decrypted Value for rule A2O1 is: 0
Decrypted Value for rule A2A2 is: 0
Decrypted Value for rule B1O1 is: 1
Decrypted Value for rule B1B1 is: 0
Decrypted Value for rule A2B1 is: 0
Decrypted Value for rule A1O1 is: 0
Decrypted Value for rule O1O3 is: 0
Decrypted Value for rule A1A1 is: 0
Decrypted Value for rule A1O3 is: 0
Decrypted Value for rule O3O3 is: 0
Decrypted Value for rule A1B1 is: 0
Decrypted Value for rule B1O3 is: 0
Decrypted Value for rule A1A2 is: 0
Decrypted Value for rule A2O3 is: 0
ABO rules are executed in 1 secs!
6.732
ABO-
2
11.976 9 1549
Decrypted Value for Haplotype O is: 1
Decrypted Value for Haplotype B is: 0
Decrypted Value for Haplotype T is: 1
Decrypted Value for EHtype O is: 1
Decrypted Value for EHtype B is: 0
Decrypted Value for EHtype T is: 0
Decrypted Value for PhHtype O is: 0
Decrypted Value for PhHtype TTT is: 0
Decrypted Value for PhHtype TTO is: 1
Decrypted Value for PhHtype BBB is: 0
Decrypted Value for PhHtype BBO is: 1
Decrypted Value for PhHtype TB is: 0
Decrypted Value for PhenoGeno O is: 1
Decrypted Value for PhenoGeno B is: 0
Decrypted Value for PhenoGeno T is: 0
Decrypted Value for PhenoGeno TB is: 0
ABO rules are executed in 5 secs!
1574
Note: Where, KG - Key Generation, E - Encryption, D- Decryption, Eval-Evaluation,
TE-Total Execution, EHtype - Extended Haplotype, PhHtype - Phenotype Haplotype,
PhenoGeno - Phenotype Genotype.
tion method and calculate average value of all pa-
tients’ results, and get the same performance results,
which are presented in both Tables 3, and 4 respec-
tively. Table 3 also presents size of original dataset,
ciphertext dataset, public key, and private key. Dis-
cussion of choosing value of “L” is presented in Sub-
section 6.2.3.
Table 4 presents the sequential computation costs
of ABO-1, and ABO-2 running on genomic datasets.
We calculate computation cost by measuring CPU
timing during key generation, encryption, ABO rule
evaluation, and decryption in seconds, and put total
execution time in last column of Table 4. In “De-
cryption (D)” column, “1” denotes presence of blood
group type and “0” denotes blood group type is not
present. These analysis are performed for all 2504
patients. We have performed all the test in 20 times
and take average value.
6.2.3 Multiplicative Depth “L”
HElib parameter “L” is considered to find the multi-
plicative depth in ABO computations. We use func-
tion “multiplyBy()” for multiplication in both ABO
rules because “multiplyBy()” is equivalent to “∗ =”
followed by a relinearization (Halevi and Shoup,
2013). Relinearization allows us to perform mul-
tiple multiplications efficiently. The relinearization
operation ensures that all ciphertext parts handle
the point to either the constant 1 or a base secret-
key (Halevi and Shoup, 2013). In HElib library,
authors have defined a higher-level method “void
Ctxt::multiplyBy(const Ctxt& other)”. This method
multiplies two ciphertexts, it begins by removing
primes from the two arguments down to a level where
the rounding-error from modulus-switching is the
dominating noise term, then it calls the low-level rou-
tine to compute the tensor product, and finally it calls
the relinearize method to get back a canonical ci-
phertext. To see the level of resulting ciphertexts
using “findBaseLevel()” from HElib library (Halevi,
2013). Afterwards we decrease initial L parameter by
“findBaseLevel()-1”. In this way resulting ciphertexts
level with new L parameter will be 1 and decryption
will work. The following equation describes the lat-
tice dimension n that is necessary to evaluate deep-L
circuits correctly with guarantee of k-bits security,
n >
(L(logn + 23) − 8.5)(k + 110)
7.2
(3)
In ABO-1 rule, the number of multiplication is 7.
So, we need level atleast 2
3
= 8 that is L = 3. If we take
L = 3, the base level will be 3-1 = 2 (Halevi, 2013),
which is a low depth level for 7 number of multiplica-
tion. If we take L = 4 (non prime number) then HElib
library automatically uses level 3, and gets base level
2 again. So, we use L = 5, and get base level 4. For
ABO-1, the value of L is L ≥ 5. Similarly, the value
of L for ABO-2 is L ≥ 15.
7 CONCLUSION
In this paper, we design an entire secure framework
for genomic datasets processing leveraging on pub-
ICISSP 2017 - 3rd International Conference on Information Systems Security and Privacy
332

lic cloud. The model protects not only genomic se-
quences but also the intermediate and final compu-
tation results when processing on public cloud. We
evaluate our proposed framework through intensive
experiments using real genomic datasets. This study
assessed the steps required for deployment of privacy-
preserving genetic testing in personalized medicine
scenario. We test the applicability of homomor-
phic encryption techniques for genetic testing with
ABO rules. This includes protection of the genomic
datasets itself and the possibility to conduct various
operations such as ABO analysis within an encrypted
environment. The testing results have proven that HE-
lib performance is close to be practical in genomic
datasets evaluation.
Our next step is to enable transciphering (Canteaut
et al., 2016) within our architecture in order to en-
hance the storage efficiency of the genomic datasets.
ACKNOWLEDGEMENT
We thank Dr. Oana Stan for fruitful discussions.
REFERENCES
Ayday, E., Cristofaro, E. D., Hubaux, J., and Tsudik, G.
(2013a). The chills and thrills of whole genome se-
quencing. ISSN: 0018-9162.
Ayday, E., Raisaro, J., and Hubaux, J.-P. (2013b). Personal
use of the genomic data: Privacy vs. storage cost. In
IEEE GLOBECOM, pages 2723–2729.
Ayday, E., Raisaro, J. L., Hengartner, U., Molyneaux, A.,
and Hubaux, J. P. (2014). Privacy-preserving process-
ing of raw genomic data. In DPM, pages 133–147.
Brakerski, Z., Gentry, C., and Vaikuntanathan, V. (2012).
(leveled) Fully homomorphic encryption without
bootstrapping. In ITCS ’12, pages 309–325.
Canteaut, A., Carpov, S., Fontaine, C., Lepoint, T., Naya-
Plasencia, M., and P. Paillier, R. (2016). Stream ci-
phers: A practical solution for efficient homomorphic-
ciphertext compression. In FSE 2016, pages 313–333.
Gentry, C., Halevi, S., and Vaikuntanathan, V. (2010). A
simple BGN-type cryptosystem from LWE. In EU-
ROCRYPT’2010, pages 506–522.
Halevi, S. (2013). HElib: An implementation of homomor-
phic encryption. https://github.com/shaih/HElib.
Halevi, S. and Shoup, V. (2013). Design and implementa-
tion of a homomorphic encryption library.
Halevi, S. and Shoup, V. (2014). Algorithms in helib. In
Cryptology-CRYPTO 2014, pages 554–571.
Jiang, X., Zhao, Y., Wang, X., Malin, B., and et al. (2014).
A community assessment of privacy preserving tech-
niques for human genomes. BMC Medical Informatics
and Decision Making, 14(1):S1.
Lauter, K., L
´
opez-Alt, A., and Naehrig, M. (2015). Private
computation on encrypted genomic data. Technical
report, Progress in Cryptology - LATINCRYPT 2014.
Lepoint, T. and Naehrig, M. (2014). A comparison of the
homomorphic encryption schemes FV and YASHE.
Cryptology ePrint Archive: Report 2014/062.
Naveed, M., Ayday, E., Clayton, E. W., Fellay, J., Gunter,
C. A., and et al. (2015). Privacy in the genomic era.
ACM Computing Surveys (CSUR), 48(1):Article No.
6.
Nogoorani, S. D. and Jalili, R. (2016). TIRIAC: A trust-
driven risk-aware access control framework for grid
environments. Future Generation Computer Systems,
55(C):238–254.
Q.Yaseen, Althebyan, Q., and Jararweh, Y. (2013). PEP-
side caching: An insider threat port. In IEEE 14th
International Conference on IRI, pages 137–144.
Smart, N. P. and Vercauteren, F. (2014). Fully homomor-
phic simd operations. Designs, Codes and Cryptogra-
phy, 71(1):57–81.
Zhang, X., Liu, C., Nepal, S., and Chen, J. (2013). An
efficient quasi-identifier index based approach for pri-
vacy preservation over incremental data sets on cloud.
Journal of Computer and System Sciences, 79(5):542–
555.
Zhou, H. and Wornell, G. (2014). Efficient homomorphic
encryption on integer vectors and its applications. In
IEEE ITA Workshop, pages 1–9.
Towards Confidentiality-strengthened Personalized Genomic Medicine Embedding Homomorphic Cryptography
333
