
Identifying Serendipitous Drug Usages in Patient Forum Data
A Feasibility Study
Boshu Ru
1
, Charles Warner-Hillard
2
, Yong Ge
3
and Lixia Yao
4,1
1
Department of Software and Information Systems, University of North Carolina at Charlotte, Charlotte, NC, U.S.A.
2
Department of Public Health Sciences, University of North Carolina at Charlotte, Charlotte, NC, U.S.A.
3
Department of Computer Science, University of North Carolina at Charlotte, Charlotte, NC, U.S.A.
4
Department of Health Sciences Research, Mayo Clinic, Rochester, MN, U.S.A.
Keywords: Social Media, Drug Repositioning, Machine Learning, Patient-Reported Outcomes.
Abstract: Drug repositioning reduces safety risk and development cost, compared to developing new drugs.
Computational approaches have examined biological, chemical, literature, and electronic health record data
for systematic drug repositioning. In this work, we built an entire computational pipeline to investigate the
feasibility of mining a new data source – the fast-growing online patient forum data for identifying and
verifying drug-repositioning hypotheses. We curated a gold-standard dataset based on filtered drug reviews
from WebMD. Among 15,714 sentences, 447 mentioned novel desirable drug usages that were not listed as
known drug indications by WebMD and thus were defined as serendipitous drug usages. We then
constructed 347 features using text-mining methods and drug knowledge. Finally we built SVM, random
forest and AdaBoost.M1 classifiers and evaluated their classification performance. Our best model achieved
an AUC score of 0.937 on the independent test dataset, with precision equal to 0.811 and recall equal to
0.476. It successfully predicted serendipitous drug usages, including metformin and bupropion for obesity,
tramadol for depression and ondansetron for irritable bowel syndrome with diarrhea. Machine learning
methods make this new data source feasible for studying drug repositioning. Our future efforts include
constructing more informative features, developing more effective methods to handle imbalance data, and
verifying prediction results using other existing methods.
1 INTRODUCTION
Drug repositioning, also known as drug repurposing,
is the identification of novel indications for
marketed drugs and drugs in the late-stage
development (Dudley et al., 2011). A well-known
example is sildenafil, which was originally
developed to treat angina in clinical trial. However,
after failure, it was resurrected to treat erectile
dysfunction (Ashburn and Thor, 2004). Another
example is the repositioning of duloxetine from
depression to stress urinary incontinence, which was
irresponsive to many drug therapies at that time
(Ashburn and Thor, 2004). These successful stories
demonstrated advantages of drug repositioning over
new drug discovery and development. Repositioned
drugs have a better safety profile than compounds in
the early discovery and development stage, as they
have already passed several preclinical tests in
animal models and safety tests on human volunteers
in the Phase I clinical trials. Thus the time and cost
of early drug discovery and development can be
saved, making repositioned drugs more available to
the patients of currently not properly treated diseases
and more cost-efficient to pharmaceutical companies
(Yao et al., 2011). Despite some potential
intellectual property issues, drug repositioning
carries the promise of significant societal benefits
and has attracted broad interests from the biomedical
community in the past decade.
Traditionally, drug-repositioning opportunities
were discovered by serendipity. In the case of
sildenafil, the clinical team was inspired with the
new repositioning idea when they found that some
patients enrolled in the original trial for angina were
reluctant to return the medicine due to the desirable
side effect (Shandrow, 2016). Various computational
methods have been developed to systematically
explore more drug-repositioning opportunities. One
common strategy is to mine chemical, biological, or
106
Ru B., Warner-Hillard C., Ge Y. and Yao L.
Identifying Serendipitous Drug Usages in Patient Forum Data - A Feasibility Study.
DOI: 10.5220/0006145201060118
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 106-118
ISBN: 978-989-758-213-4
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

clinical data for drug similarity, disease comorbidity,
or drug-disease associations that imply repositioning
opportunities (Dudley et al., 2011, Andronis et al.,
2011). For instance, Keiser et al. (2009) compared
chemical structure similarities among 3,665 drugs
and 1,400 protein targets to discover unanticipated
drug-target associations and implicated the potential
role of Fabahistin, an allergy drug, in treating
Alzheimer’s disease. Sanseau et al. (2012)
investigated data from genome-wide association
studies to systematically identify alternative
indications for existing drugs and suggested
repositioning denosumab, which was approved for
osteoporosis, for Crohn's disease. Hu and Agarwal
(2009) created a drug-disease network by mining the
gene-expression profiles in GEO datasbase and the
Connectivity Map project. By analyzing topological
characteristics of this network, they inferred the
effects of cancer and AIDS drugs for Huntington's
disease. Wren et al. (2004) constructed a network of
biomedical entities including genes,
diseases/phenotypes, and chemical compounds from
MEDLINE (U.S. National Library of Medicine,
2016a), and computationally identified novel
relationships between those biomedical entities in
scientific publications. One such relationship they
found and validated in the rodent model was
between chlorpromazine and cardiac hypertrophy.
Gottlieb et al. (2011) designed an algorithm called
PREDICT, to discover novel drug-disease
associations from OMIM, DrugBank, DailyMed,
and Drugs.com. Their algorithm predicted 27% of
drug-disease associations in clinical trials registered
with clinicaltrial.gov. Although these computational
methods have demonstrated their promise, they often
face the issue of high false positive rates (Dudley et
al., 2011, Shim and Liu, 2014). One primary reason
is sharing similar chemical structures or co-
occurring in the same publication does not always
imply medical relevance. Also, ignoring the context
(e.g., whether the similarity or validation is observed
in experiments on molecular, cell line, or animal
models) might impact their capability to be
translated to human beings.
More recently, researchers began to verify some
drug-repositioning hypotheses using the Electronic
Health Record (EHR) data. For example, Khatri et
al. (2013) retrospectively analyzed the EHR of 2,515
renal transplant patients at the University Hospitals
Leuven to confirm the beneficial effects of
atorvastatin on graft survival. Xu et al. (2014)
verified that metformin, a common drug for type 2
diabetes, is associated with improved cancer survival
rate by analyzing the patients’ EHR data from
Vanderbilt University Medical Center and Mayo
Clinic. These proof-of-concept studies also
witnessed several limitations, due to the nature of
EHR data: (1) EHR systems do not record the causal
relationships between events (e.g., drugs and side
effects) as they are mostly designed for clinical
operation and patient management instead of
research. Whether a statistical association is causal
needs to be verified through temporal analysis with a
lot of assumptions. Therefore, the models become
disease and/or drug specific and remain difficult to
generalize and automate in large scale. (2) A
significant amount of valuable information, such as
the description of medication outcomes, is stored in
clinicians’ notes in free-text format (Yao et al.,
2011). Mining these notes requires advanced natural
language processing techniques and presents patient
privacy issues. (3) In the US, data from a single
provider's EHR system only provide an incomplete
piece of patient care (Xu et al., 2014). Integrating
EHR data from multiple providers may be a
solution, but currently encounters legal and technical
challenges, as discussed in depth by Jensen et al.
(2012). Due to these limitations, neither EHR, nor
any of scientific literature, biological, and chemical
data alone appear sufficient for drug repositioning
research. We need to identify additional data sources
that contain patient medication history and
outcomes, as well as develop advanced data
integration methods to identify synergistic signals.
In the last decade or so, another type of patient
data has increased exponentially in volume with the
emergence of smart phones and social media
websites. People today not only post their travel
pictures but also share and discuss their experiences
with diseases and drugs in patient forums and social
media websites, such as WebMD, PatientsLikeMe,
Twitter, and YouTube (Ru et al., 2015). Such data
directly describes drug-disease associations in real
human patients and bypasses the translational hurdle
from cell-line or animal model to human, thus has
led to increased research interests. For example,
Yang et al. (2012) detected adverse drug reaction
(ADR) signals from drug related discussions in the
MedHelp forum by using an ADR lexicon created
from the Consumer Health Vocabulary. Yates and
Goharian (2013) extracted ADR in the breast cancer
drug reviews on askpatient.com, drugs.com, and
drugratingz.com using a ADR synonym list
generated from the United Medical Language
System (UMLS) specifically for breast cancer.
Rather than collecting existing social media
discussions, Knezevic et al. (2011) created a
Facebook group for people to report their ADR
outcomes and found social media a highly sensitive
Identifying Serendipitous Drug Usages in Patient Forum Data - A Feasibility Study
107
instrument for ADR reporting . Powell et al. (2016)
investigated the MedDRA Preferred Terms that
appeared on Twitter and Facebook and found 26%
of the posts contained useful information for post-
marketing drug safety surveillance.
In this work, we expand current social media
mining research that is primarily ADR focused to
the discovery of serendipitous drug usages, which
can suggest potentially new drug repositioning
hypotheses. We build a computational pipeline
based on machine learning methods to capture the
serendipitous drug usages on the patient forum
published by WebMD, which was reported in a
previous study (Ru et al., 2015) to have high-quality
patient reported medication outcomes data.
However, this is an extremely difficult machine
learning task because: (1) User comments on patient
forum are unstructured and informal human
language prevalent with typographic errors and chat
slangs. It is unclear how to construct meaningful
features with prediction power; (2) the mentioning
of serendipitous drug usages by nature is very rare.
Based on our experience with the drug reviews on
WebMD, the chance of finding a serendipitous drug
usage in user posts is less than 3% (See Methods).
Therefore, we caution the audience that our
objective in this work is not to build a perfect
pipeline or a high performance classifier, but to
perform a feasibility check and identify major
technical hurdles in the entire workflow. We plan to
direct our systems engineering efforts towards
improving the performance of those bottleneck
modules as the next step.
2 METHODS
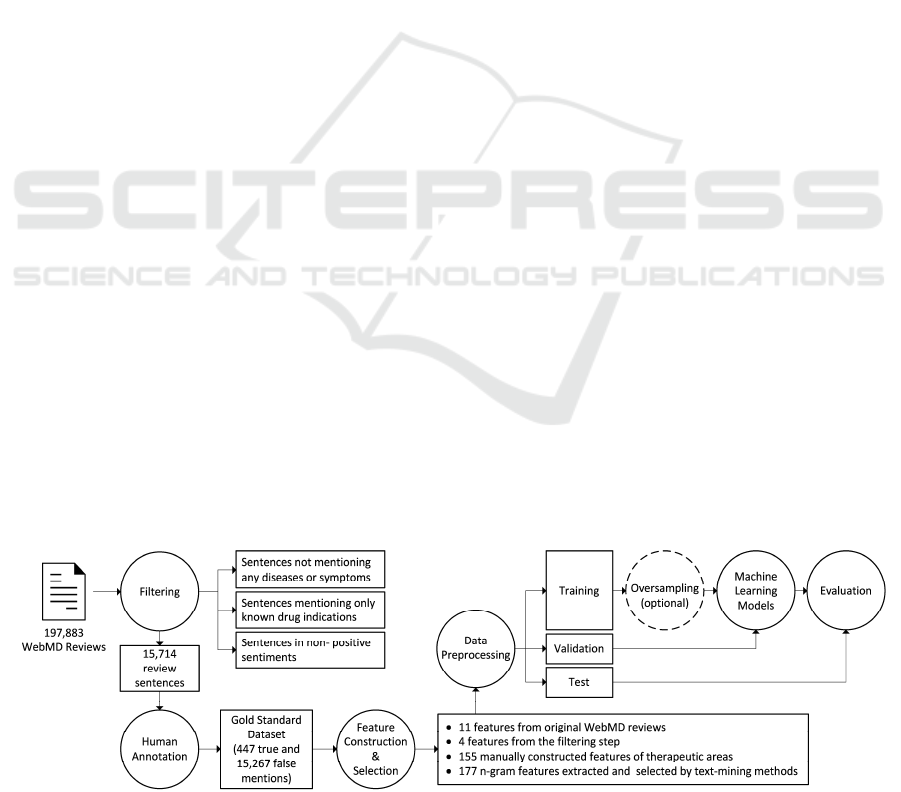
In this feasibility study, we built the entire
computational pipeline using standard tools and
applications, to identify serendipitous drug usages in
patient forum data, which includes data collection,
data filtering, human annotation, feature
construction and selection, data preprocessing,
machine learning model training and evaluation, as
illustrated in Figure 1. Each module is further
described below.
2.1 Data Collection
We started by collecting drug reviews posted by
anonymous users on the patient forum hosted by
WebMD. WebMD is a reputable health care website
that exchanges disease and treatment information
among patients and healthcare providers. In its
patient forum, after filling the basic demographic
information including gender and age group, users
are allowed to rate drugs in terms of effectiveness,
ease of use, overall satisfaction, and post additional
comments about their medication experience (See
Figure 2). We chose it based on two considerations:
(1) With over 13 years’ history of operation and on
average over 150 million unique visits per month,
WebMD contains a large volume of drug reviews
that is highly desirable for conducting systematic
studies. (2) The quality of drug reviews was reported
to be superior to many other social media platforms
in a previous study (Ru et al., 2015). Spam reviews,
commercial advertisements, or information
irrelevant to drugs or diseases are rare, probably
thanks to their forum modulators. We downloaded a
total number of 197,883 user reviews on 5,351 drugs
by the date of March 29, 2015. Then, we used
Stanford CoreNLP (Manning et al., 2014) to break
down each free-text comment into sentences, which
is the standard unit for natural language processing
and text mining analysis.
2.2 Gold Standard Dataset for
Serendipitous Drug Usages
In machine learning and statistics, gold standard, or
accurately classified ground truth data is highly
desirable, but always difficult to obtain for
Figure 1: A workflow to identify serendipitous drug usages in patient forum data.
HEALTHINF 2017 - 10th International Conference on Health Informatics
108

supervised learning tasks. For identifying
serendipitous drug usages, it would be ideal if a
database of drug usages approved globally or
customarily used off-label were readily available as
the benchmark for known drug usages. The
professional team at WebMD has published
monographs to introduce each drug, including
information on drug use, side effects, interactions,
overdose, etc. We thus used such data as the
benchmark for known drug usages in this work. We
assume a drug use is serendipitous if the user
mentioned improvement of his or her condition or
symptom that was not listed in the drug's known
indications according to WebMD (See the examples
in Figure 2). Otherwise, we set the mentioned drug
use to be non-serendipitous. Below we explain in
more details how we applied this principal to semi-
automatically prepare our gold standard dataset for
serendipitous drug usages.
2.3 Data Filtering
Three filters were designed to reduce the number of
drug review sentences to a number more manageable
for human annotation. Firstly, we identified and
removed review sentences that did not mention any
disease or symptom at all, because these sentences
have no chance to be related to serendipitous drug
usages. To do this, we selected the UMLS concepts
in English and with the semantic types equal to
Disease or Syndrome, Finding, Injury or Poisoning,
Mental or Behavioral Dysfunction, Neoplastic
Process, or Sign or Symptom and used them to
approximate medical concepts that could be related
to serendipitous drug usages. We then used
MetaMap (Aronson and Lang, 2010) to identify
these medical concepts in each review sentence.
Next, for sentences that did mention any of those
concepts, we used SNOMED CT (U.S. National
Library of Medicine, 2016b) to determine whether
the mentioned concept is semantically identical or
similar to the drug's known indications listed on
WebMD. Mathematically SNOMED CT is a
directed acrylic graph model for medical
terminology. Medical concepts are connected by
defined relationships, such as is-a, associated with,
and due to. The semantic similarity between two
concepts was usually measured by the length of the
shortest path between them in the graph (Pedersen et
al., 2007, Shah and Musen, 2008). If the medical
concept mentioned in a review sentence was more
than three steps away from the known indications of
the drug, we assumed the mentioned medical
concept was more likely to be an unanticipated
outcome for the drug and kept the sentence in the
dataset for the third filter. Otherwise, we excluded
the sentence from further evaluation, as it was more
likely to be related to the drug’s known usage rather
than serendipitous usage we were looking for. In the
third step, we used the sentiment analysis tool,
Deeply Moving (Socher et al., 2013) offered by the
Stanford Natural Language Processing Group to
assess the sentiment of each sentence where
unanticipated medical concept occurred. We filtered
out all sentences with Very Negative, Negative, or
Neutral sentiment and only kept those with Positive
or Very Positive sentiments because serendipitous
drug usages are unexpected but desirable outcomes
to patients. Negative sentiment is more likely to be
associated with undesirable side effects or potential
drug safety concerns. After these three filtering
steps, 15,714 drug review sentences remained for
further human annotation.
2.4 Human Annotation
One public health professional and one health
informatics professional with master degrees,
independently reviewed the 15,714 sentences and
annotated whether each sentence was a true mention
of serendipitous drug usage based on the benchmark
Figure 2: Examples of serendipitous drug usage mention on WebMD. In the example on the left, a patient reported that his
irritable bowel syndrome (IBS) symptoms were alleviated when taking sulfasalazine to treat rheumatoid arthritis. In the
example on the right, an asthma patient taking prednisone reported the improvement of her eczema.
Identifying Serendipitous Drug Usages in Patient Forum Data - A Feasibility Study
109
dataset of known drug usages defined by WebMD.
That is, they labeled a drug use to be serendipitous if
the user mentioned an improved condition or
symptom that was not listed in the drug's known
indications according to WebMD. Otherwise, they
assigned the mentioned drug use to be non-
serendipitous. In case that the annotators did not
agree with each other, they discussed and assigned a
final label together. Six months later, the two
professionals reviewed their annotation again to
avoid possible human errors. In total, 447 or 2.8% of
sentences were annotated to contain true
serendipitous drug usage mentions, covering 97
drugs and 183 serendipitous drug usages. The rest
15,267 sentences were annotated to contain no
serendipitous drug usage mentions. This dataset was
used throughout the study as the gold standard
dataset to train and evaluate various machine
learning models.
2.5 Feature Construction and Selection
Feature construction and selection is an important
part of data mining analysis, in which the data is
processed and presented in a way understandable by
machine learning algorithms. The original drug
reviews downloaded from WebMD website come
with 11 features, including patients’ ratings of drug
effectiveness, ease of use, overall satisfaction, and
the number of people who thought the review is
helpful (See Table 1).
In the data-filtering step, we created four more
features, which are (1) whether the sentence contains
negation, (2) the UMLS semantic types of
mentioned medical concepts; (3) the SNOMED CT-
based semantic distance between a drug's known
indication and the medical concept the user
mentioned in a review sentence; (4) the sentiment
score of the review sentence.
Prior knowledge in drug discovery and
development also tells that some therapeutic areas,
such as neurological disorders, bacteria infection,
and cancers are more likely to have “dirty” drugs,
which bind to many different molecular targets in
human body, and tend to have a wide range of
effects (Yao and Rzhetsky, 2008, Frantz, 2005,
Pleyer and Greil, 2015). Therefore, drugs used in
those therapeutic areas have higher chance to be
repositioned. We manually selected 155 drug usages
from those therapeutic areas and used them as binary
features, which hopefully capture useful information
and improve machine learning predictions of
serendipitous drug usages.
We also adopted a commonly used text-mining
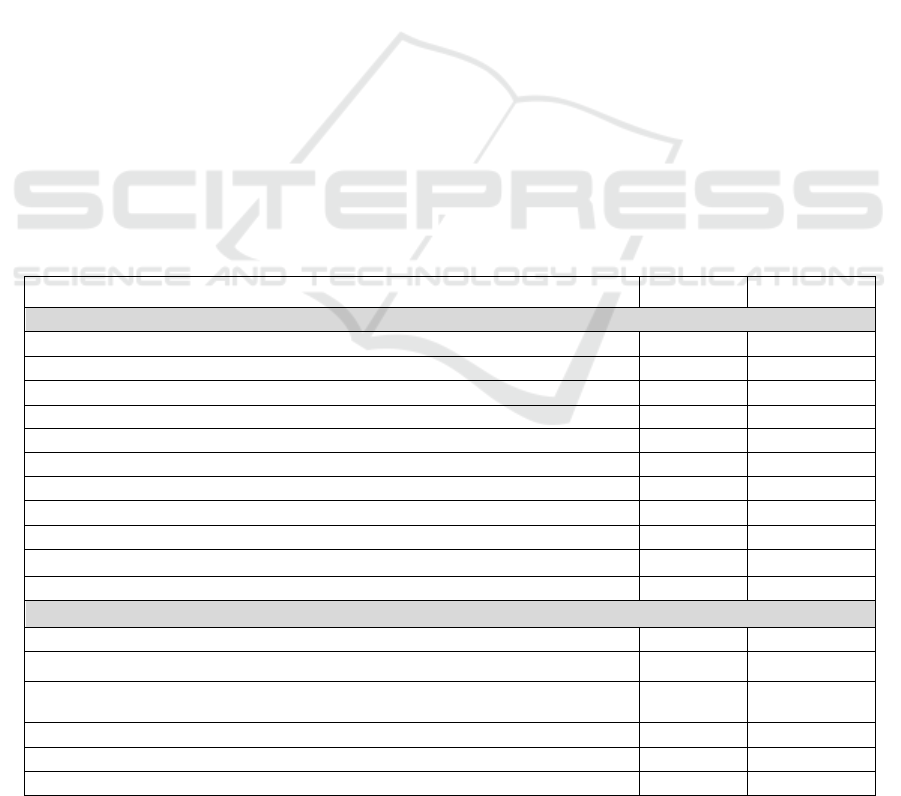
Table 1: List of the features constructed for the annotated datasets.
Name Data Type Source
Original Features obtained from the Patient Forum
User rating of effectiveness Numerical WebMD
User rating of ease of use Numerical WebMD
User rating of overall satisfaction Numerical WebMD
Number of users who felt the review was helpful Numerical WebMD
Number of reviews for the drug Numerical WebMD
The day of review Categorical WebMD
The hour of review Categorical WebMD
User's role (e.g., Patient, Caregiver) Categorical WebMD
User's gender Categorical WebMD
User’s age group Categorical WebMD
The time on the drug (e.g., less than 1 month, 1 to 6 months, 6 months to 1 year) Categorical WebMD
Additional Features
Whether the sentence contains negation Binary MetaMap
Semantic types of medical concepts mentioned in the sentence Categorical MetaMap
Semantic distance between the mentioned medical concept and the drug’s known
indications in SNOMED CT
Numerical SNOMED
Sentiment score Numerical Deeply Moving
Therapeutic areas (155) Binary Self-constructed
N-grams extracted from drug review sentences (177) Binary Self-constructed
HEALTHINF 2017 - 10th International Conference on Health Informatics
110

method, n-gram (Fürnkranz, 1998), to generate more
textual features. An n-gram is a contiguous sequence
of n words from a given text and it captures the
pattern about how people use word combination in
their communication. We used the tm package in R
(Feinerer and Hornik, 2012) to do this. After the
steps of punctuation and stop words removal, word
stemming, and rare words pruning, we extracted
3,264 unigrams, 10,064 bigrams, and 5,058 trigrams.
For each n-gram, we calculated the information gain
(Michalski et al., 2013) to assess its differentiating
power between true and false classes in Weka (Hall
et al., 2009). We excluded n-grams whose
information gain equaled zero and kept 177 n-grams
with positive information gain (namely 64 unigrams,
73 bigrams, and 40 trigrams) as additional textual
features. In total, 347 features were constructed for
the machine learning classification, as summarized
in Table 1.
2.6 Data Preprocessing
We normalized the data by linearly re-scaling all
numerical features to the range of [-1, 1]. Such
processing is necessary for support vector machine
(SVM) to ensure no features dominate the
classification just because of their order of
magnitude, as SVM calculates the Euclidean
distances between support vectors and the separation
hyperplane in high-dimensional space (Ali and
Smith-Miles, 2006). Then we split the 15,714
annotated sentences into training, validation, and test
datasets, according to their post dates. Sixty percent
of them, or 9,429 sentences posted between
September 18, 2007 and December 07, 2010, were
used as the training dataset to build machine
learning models. Twenty percent of the data, or
3,142 sentences posted between December 08, 2010
and October 11, 2012 were used as the validation
dataset to tune the model parameters. The remaining
20% of data, or 3,143 sentences that were posted
between October 12, 2012 and March 26, 2015,
were held as the independent test dataset. The
proportion of serendipitous drug usages in the three
datasets was between 2.0% and 3.2%. This
arrangement is essential to pick up the models that
could generalize on future and unseen data and
minimize the bias led by overfitting, as the
validation and test datasets occur temporally after
the training dataset.
2.7 Machine Learning Models
We selected three state-of-art machine learning
algorithms, namely SVM (Cortes and Vapnik,
1995), random forest (Breiman, 2001) and
AdaBoost.M1 (Freund and Schapire, 1996) to build
the prediction models. The implementation was
based on Weka (version 3.7) (Hall et al., 2009) and
LibSVM library (Chang and Lin, 2011). For SVM,
we used the radial basis function (RBF) kernel and
conducted grid search to find the optimal parameters
including C and gamma (γ). LibSVM is able to
produce both probability estimates (Wu et al., 2004)
and class labels as output. For random forest, we
empirically set the number of trees to be 500 and
iteratively searched for the optimal value for number
of features. By default the prediction gives a
probability estimate for each class. For
AdaBoost.M1, we selected the decision tree built by
C4.5 algorithm (Quinlan, 2014) as the weak learner
and obtained the optimal value for number of
iterations through iterative search. The Weka
implementation of AdaBoost.M1 only provides class
labels as prediction results. Our evaluation therefore
is based on class label predictions from all three
algorithms, without considering the probability
estimates from SVM and random forest.
As the chance of finding a serendipitous drug
usage (positive class) is rare and the vast majority of
the drug reviews posted by users do not mention any
serendipitous usages (negative class), we were
facing an imbalanced dataset problem. Therefore,
we used the oversampling technique (He and Garcia,
2009, Batuwita and Palade, 2010, Kotsiantis et al.,
2006) to generate another training dataset where the
proportion of positive class was increased from
2.8% to 20%. Afterward, we tried the same machine
learning algorithms on the oversampled training
dataset, and compared the prediction results side-by-
side with those from the original, imbalanced
training dataset.
2.8 Evaluation
We were cautious about choosing appropriate
performance evaluation metrics because of the
imbalanced dataset problem. Of commonly used
metrics, accuracy is most vulnerable to imbalanced
dataset since a model could achieve high accuracy
simply by assigning all instances into the majority
class. Instead we used a combination of three
commonly used metrics, namely precision, recall,
and area under the receiver operating characteristic
curve (also known as AUC score) (Caruana and
Niculescu-Mizil, 2004), to evaluate the performance
of various prediction models on the independent test
dataset. We also conducted 10-fold cross validation
Identifying Serendipitous Drug Usages in Patient Forum Data - A Feasibility Study
111

by combining training, validation and testing
datasets together, in order to compare our results
directly with some other drug-repositioning studies.
In addition, we manually reviewed 10% of
instances in the test dataset that were predicted to be
serendipitous drug usages and searched through the
scientific literature to check if these predictions
based purely on machine learning methods can
replicate the discoveries from biomedical scientific
community, as another verification on whether
machine learning methods alone can potentially
predict completely new serendipitous drug usages.
All our data and scripts from this work will be
made available to academic users upon request.
3 RESULTS
3.1 Parameter Tuning
We used AUC score to tune the model parameters
on the validation dataset. In case that the AUC
scores of two models were really close, we chose the
parameter and model that yielded higher precision.
This is because end users (e.g., pharmaceutical
scientist) are more sensitive to cases that were
predicted to be the under-presented, rare events,
which are serendipitous drug usages in this work,
when they evaluate the performance of any kind of
machine learning based predictive models. For SVM
models, the optimal value of gamma (γ), the width
of RBF kernel was 0.001 without oversampling and
0.1 with oversampling. The optimal value of C,
which controls the trade-off between model
complexity and ratio of misclassified instances, was
equal to 380 without oversampling and 0.1 with
oversampling. For random forest models, the
number of features decides the maximum number of
features used by each decision tree in the forest,
which was found to be 243 without oversampling
and 84 with oversampling at the best performance
on validation dataset. For AdaBoost.M1, the number
of iterations specifies how many times the weak
learner will be trained to minimize the training error.
Its optimal value equaled 36 without oversampling
and 58 with oversampling.
3.2 Performance Metrics
We evaluated the performance of six prediction
models, namely SVM, random forest and
AdaBoost.M1 with and without oversampling, on
independent test dataset. The results were
summarized in Table 2. The highest AUC score
(0.937) was achieved from the AdaBoost.M1 model,
whereas the lowest score (0.893) was from the SVM
with oversampling. On the whole, AUC scores for
all models were higher than 0.89, demonstrating the
promise of machine learning models for identifying
serendipitous drug usages from patient forums.
The precision of random forest and
AdaBoost.M1 models with and without
oversampling, and the SVM model without
oversampling were between 0.758 and 0.857, with
the highest precision achieved on the random forest
model without oversampling. However, the
precision for the SVM model with oversampling was
0.474, which was significantly lower than the other
models. The recall of all models was less than 0.50.
This means more than 50% of serendipitous usages
were not identified. Obtaining either low recall or
low precision remains a common challenge for
making predictions from extremely imbalanced
datasets like ours (He and Garcia, 2009). In many
cases, it becomes a compromise depending on the
application and the users’ need. In our experiment,
after we increased the proportion of the positive
class to 20% by oversampling, the recall of SVM
and random forest models increased slightly; but the
precision and the AUC score decreased.
Oversampling seemed ineffective on AdaBoost.M1
models. The AUC score, precision and recall for
AdaBoost.M1 with oversampling all decreased,
Table 2: Model performance in terms of precision, recall and AUC score.
Model
Test dataset 10-fold cross validation
AUC Precision Recall AUC Precision Recall
SVM 0.900 0.758 0.397 0.926 0.817 0.539
SVM - Oversampling 0.893 0.474 0.429 0.932 0.470 0.620
Random Forest 0.926 0.857 0.381 0.935 0.840 0.506
Random Forest - Oversampling 0.915 0.781 0.397 0.944 0.866 0.530
AdaBoost.M1 0.937 0.811 0.476 0.949 0.791 0.575
AdaBoost.M1 - Oversampling 0.934 0.800 0.444 0.950 0.769 0.559
HEALTHINF 2017 - 10th International Conference on Health Informatics
112

compared to the metrics on AdaBoost.M1 models
without oversampling. In the 10-fold cross
validation experiment, both recall and AUC scores
seemed to be better than what were observed on the
independent test set. Our AUC scores were close to
the same scores reported by the drug-repositioning
algorithm of PREDICT (Gottlieb et al., 2011), which
were also from a 10-fold cross validation.
3.3 Prediction Review
For the 10% of instances in the test dataset that were
predicted to be serendipitous drug usages, we
conducted a literature and clinical trial search to
provide a closer verification of our prediction
models. Table 3 summarizes the analysis. We also
presented the condensed evidences in literature
and/or clinical trial below, for each instance.
3.3.1 Metformin and Obesity
A patient reported weight loss while taking
metformin, a type 2 diabetes drug. Actually in the
past two decades, metformin's effectiveness and
safety for treating obesity in adult and child patients
have been clinically examined in dozens of clinical
trials and meta-analyses studies with promising
results (Igel et al., 2016, Desilets et al., 2008,
Paolisso et al., 1998, Peirson et al., 2014, McDonagh
et al., 2014). According to the literature review by
Igel et al. (2016), one possible explanation is that
metformin could increase the body’s insulin
sensitivity, which helps obese patients (who
typically develop resistance to insulin) to reduce
their craving for carbohydrates and to reduce the
glucose stored in their adipose tissue. Other
explanations include that metformin may enhance
energy metabolism by accelerating the
phosphorylation of the AMP-activated protein
kinase system, or it may cause appetite loss by
correcting the sensitivity and resistance of leptin.
3.3.2 Painkiller and Depression
When tramadol was taken for back pain, a patient
found it also helpful with his depression and anxiety.
Tramadol is an opioid medication, which have been
long used for the psychotherapeutic benefits
(Tenore, 2008). Tetsunaga et al. (2015) have
demonstrated tramadol's efficacy in reducing
depression levels among lower back pain patients
with depression in an 8-week clinical trial. The self-
reported depression scale of patients in the tramadol
group was 6.5 points lower than the control group.
Similarly the combinatory therapy of acetaminophen
and oxycodone, another painkiller, was reported by
Stoll and Rueter (1999) to have antidepressant effect
too.
3.3.3 Bupropion and Obesity
In the specific comment, the patient reported that
Bupropion, an anti-depressant, helped him to lose
weight. The weight loss effect of bupropion might
be attributed to increased dopamine concentration in
the brain, which leads to suppressed appetite and
reduced food intake (Greenway et al., 2010). This
serendipitous drug usage was also supported by
several clinical trials (Gadde et al., 2001, Anderson
et al., 2002, Jain et al., 2002).
3.3.4 Ondansetron and Irritable Bowel
Syndrome with Diarrhea
Ondansetron is a medication for nausea and
vomiting. Sometimes it causes the side effect of
constipation in patients. Interestingly, this patient
also had irritable bowel syndrome with diarrhea and
thus ondansetron helped to regulate that. This
serendipitous usage actually highlights the
justification of personalized medicine and has been
tested in a recent clinical trial (Garsed et al., 2014).
3.3.5 Desvenlafaxin and Lack of Energy
In the last case, anti-depressant desvenlafaxine was
reported to boost energy. Strictly speaking, lack of
energy is not a disease but a symptom. With limited
information on the patient's physical and
psychological conditions before and after
medication, it remains unclear whether the energy
boost effect was due to changes in the neural system
or was purely a natural reflection of more positive
moods after the patient took the anti-depressant
medicine. We did not find any scientific literature
discussing the energy boost effect of desvenlafaxine.
So this case could represent either a new
serendipitous drug use or a promiscuous drug usage.
Identifying Serendipitous Drug Usages in Patient Forum Data - A Feasibility Study
113
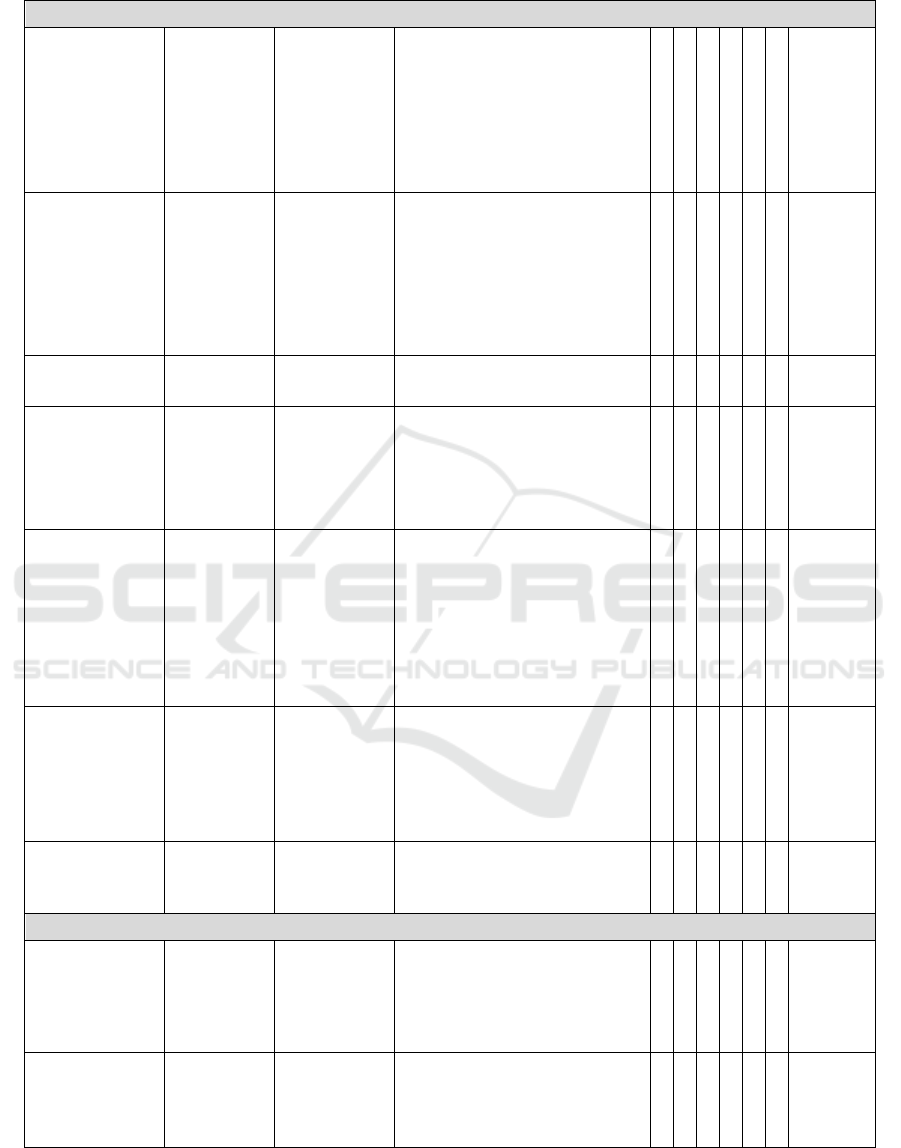
Table 3: Examples of serendipitous drug usages predicted by the models.
True positive examples
Drug
Known
indications
Serendipitous
usage
Example
SVM
SVM-Oversam
p
lin
g
RF
*
RF-Oversam
p
lin
g
*
Ada
*
Ada-Oversam
p
lin
g
*
Literature evidence
Metformin
Type 2
Diabetes
Mellitus,
Polycystic
Ovary
Syndrome,
etc.
Obesity
I feel AWFUL most of the day,
but the weight loss is great.
x x x x x x
Igel et al.
(2016),
Desilets et
al. (2008),
Paolisso et
al. (1998)
Tramadol Pain
Depression,
anxiety
It also has helped with my
depression and anxiety.
xx
x x
Tetsunaga et
al. (2015)
Acetaminophen
& oxycodone
Pain Depression
While taking for pain I have
also found it relieves my major
depression and actually gives
me the energy and a clear mind
to do things.
xxx x
Stoll and
Rueter
(1999)
Bupropion
Depression,
attention
deficit &
hyperactivity
disorder
Obesity
I had energy and experienced
needed weight loss and was
very pleased, as I did not do
well on SSRI or SNRIs.
xx
x x x
Greenway et
al. (2010),
Gadde et al.
(2001),
Anderson et
al. (2002),
Jain et al.
(2002)
Ondansetron Vomiting
Irritable
bowel
syndrome
with diarrhea
A lot of people have trouble
with the constipation that comes
with it, but since I have IBS-D
(irritable bowel syndrome with
diarrhea), it has actually
regulated me .
x x
Garsed et al.
(2014)
Desvenlafaxine Depression
Lack of
energy
I have had a very positive mood
and energy change, while also
experiencing much less anxiety.
x x x x x
False positive examples
5-HTP
Anxiety,
depression
Thyroid
Diseases,
Obesity
i have Hoshimitos thyroid
disease
**
and keeping stress
levels down is extremely
important for many reasons but
also for weight loss.
x x
Cyclobenzaprine
Muscle
spasm
Pain
While taking this medication for
neck stiffness and pain; I
discovered it also helped with
other muscle spasms.
x
*
RF stands for random forest. Ada stands for AdaBoost.M1. "x" indicates the model recognized the example as a
serendipitous usage.
**
Hoshimitos thyroid disease was a typo. The correct spelling should be Hashimoto's Thyroiditis.
HEALTHINF 2017 - 10th International Conference on Health Informatics
114

3.3.6 False Positive Predictions
Besides the true positive examples, we also found
two cases where some of our models made false
positive predictions due to difficult language
expression and terminology flaw. The first example
is 5-HTP, an over-the-counter drug for anxiety and
depression. One patient commented that stress relief
brought by this drug was important to her
Hashimito's thyroid disease and weight loss.
Although Hashimoto's disease and weight loss were
mentioned, the patient did not imply the 5-HTP can
treat Hashimoto’s disease or control weight. But
SVM and random forest models with over-sampling
became confused by the subtle semantic difference.
In the second case, a patient taking cyclobenzaprine
for neck stiffness and pain said the drug also helped
with other muscle spasms. Pain, neck stiffness and
muscle spasms are really close medical concepts.
We found that this false positive prediction was
actually due to imperfect terminology mapping.
4 DISCUSSION
In this very first effort to identify serendipitous drug
usages from online patient forum, we designed an
entire computational pipeline. This feasibility study
enabled us to thoroughly examine the technical
hurdles in the entire workflow and answer the
question if patient-reported medication outcome data
on social media is worthwhile to explore for drug
repositioning research. The best-performing model
was built from AdaBoost.M1 method without
oversampling, which had precision equal to 0.811,
recall equal to 0.476 and AUC score equal to 0.937
on independent test data. The 10-fold cross
validation results are also comparable to existing
drug-repositioning method (Gottlieb et al., 2011).
Therefore our confidence in applying machine
learning methods to identify serendipitous drug
usages from online patient forum data is increased.
More specifically we have addressed the following
tasks in this work:
Previously, there was no curated social media
dataset available for the purpose of identifying
serendipitous drug usages. We spent a considerable
amount of time and effort to collect, filter and
annotate 15,714 drug review sentences from the
WebMD patient forum site. Two health
professionals at master level annotated all the
sentences independently and discussed on cases
when disagreement occurred. They repeated this
process six months later. If more resource available,
we would like to recruit a larger group of
professionals to curate a larger and more reliable
gold standard dataset. But the current annotated
dataset is comprehensive enough for this work, as it
covers not only easy instances, but also challenging
ones for machine learning prediction, as shown in
Table 3.
In addition, the drug reviews posted on patient
forum are unstructured and informal human
language prevalent with typographic errors and chat
slangs, which need to be transformed to a
representation of feature vectors before machine
learning algorithms could comprehend. We used
patients’ demographic information, ratings of drug
effectiveness, ease of use, and overall satisfaction
from the patient forum. We calculated negation,
semantic similarity between the unexpected
medication outcome mentioned in a review sentence
and the known drug indications based on SNOMED
CT, and sentiment score of the review sentence. We
also leveraged our known knowledge on dirty drugs,
and extracted informative n-gram features based on
information gain. The results from this feasibility
study showed that these features are useful to predict
serendipitous drug usages. For example, dirty drugs
for neurological conditions did show up
predominantly in the results. But these features
seemed not sufficient to predict all serendipitous
drug usages correctly. As shown in the false positive
examples of Table 3, the n-grams such as also, also
help, and also for were often associated with true
serendipitous drug usages, but could occur in false
positive cases too. Current medical terminology
mapping tools (i.e., MetaMap) could be the
performance-limiting step in cases like pain and
muscle spasm, despite the close connection of these
two concepts from the perspective of medicine. We
will explore more sophisticated methods such as
DNorm (Leaman et al., 2013), as well as additional
methods of semantic similarity calculation as shown
in (Pedersen et al., 2007, Sánchez et al., 2012) in
future.
Thirdly, the data are extremely imbalanced
between two classes (2.8% vs. 97.2%) because
serendipitous drug usages are rare events by nature.
Such imbalance inevitably impedes the performance
of machine learning algorithms. We tried to increase
the proportion of serendipitous usages in the training
dataset to 20%, using the random oversampling
method (He and Garcia, 2009). We have also tried
two other methods, namely synthetic minority over-
sampling technique (Chawla et al., 2002) and under-
sampling (Kotsiantis et al., 2006), but their
performance was inferior to that of random
Identifying Serendipitous Drug Usages in Patient Forum Data - A Feasibility Study
115

oversampling and not shown here. More robust
machine learning algorithms that are less sensitive to
imbalanced data or robust sampling methods will be
desirable to further improve serendipitous drug
usage predictions.
Last but not least, we acknowledge that as an
emerging data source, online patient forums have
limitations too. Many patients who write drug
reviews online lack of basic medical knowledge.
Their description of the medication experience can
be ambiguous, hyperbolic or inaccurate. Also
important contextual information, such as co-
prescribed drugs, may be missed in the review.
Without a comparison between an experiment group
and a control group, serendipitous drug usages
extracted from patient forums need to be further
verified for drug repositioning opportunities by
integrating with existing data sources, such as EHR
and scientific literature.
5 CONCLUSIONS
Drug repositioning is an important but not yet fully
utilized strategy to improve the cost-effectiveness of
medicine and to reduce the development time. The
dawn of social media brings large volumes of
patient-reported medication outcome data, and thus
creates an urgent need to examine it for the purpose
of drug repositioning. In this work, we collected,
filtered, and annotated drug review comments posted
on WebMD patient forum. We built an entire
computational pipeline based state-of-art machine
learning and text mining methods to mine
serendipitous drug usages. Our models achieved
AUC scores that are comparable to existing drug
repositioning methods. Most instances that were
predicted to be serendipitous drug usages are also
supported by scientific literature. So machine
learning approaches seem feasible to address this
problem of looking for a needle in the haystack.
More of our future efforts will be directed to develop
more informative features, improve disease mapping
accuracy, handle imbalanced data, and integrate
findings from social media with other data sources,
in order to build really functional drug-repositioning
applications.
REFERENCES
Ali, S. & Smith-Miles, K. A. Improved support vector
machine generalization using normalized input space.
In: Proceedings of the 19th Australasian Joint
Conference on Artificial Intelligence, 2006 Hobart,
Australia. Springer, 362-371.
Anderson, J. W., Greenway, F. L., Fujioka, K., Gadde, K.
M., Mckenney, J. & O'neil, P. M. 2002. Bupropion SR
Enhances Weight Loss: A 48-Week Double-Blind,
Placebo-Controlled Trial. Obesity Research, 10, 633-
641.
Andronis, C., Sharma, A., Virvilis, V., Deftereos, S. &
Persidis, A. 2011. Literature mining, ontologies and
information visualization for drug repurposing.
Briefings in Bioinformatics, 12, 357-368.
Aronson, A. R. & Lang, F.-M. 2010. An overview of
MetaMap: historical perspective and recent advances.
Journal of the American Medical Informatics
Association, 17, 229-236.
Ashburn, T. T. & Thor, K. B. 2004. Drug repositioning:
identifying and developing new uses for existing
drugs. Nature Review Drug Discovery, 3, 673-683.
Batuwita, R. & Palade, V. Efficient resampling methods
for training support vector machines with imbalanced
datasets. In: Proceedings of the International Joint
Conference on Neural Networks (IJCNN), 2010
Barcelona, Spain. IEEE, 1-8.
Breiman, L. 2001. Random forests. Machine Learning, 45,
5-32.
Caruana, R. & Niculescu-Mizil, A. Data mining in metric
space: an empirical analysis of supervised learning
performance criteria. In: Proceedings of the 10th ACM
SIGKDD International Conference on Knowledge
Discovery and Data Mining, 2004 Seattle, WA, USA.
ACM, 69-78.
Chang, C. & Lin, C. 2011. LIBSVM: a library for support
vector machines. ACM Transactions on Intelligent
Systems and Technology (TIST), 2, 27.
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer,
W. P. 2002. SMOTE: synthetic minority over-
sampling technique. Journal of Artificial Intelligence
Research, 16, 321-357.
Cortes, C. & Vapnik, V. 1995. Support-vector networks.
Machine Learning, 20, 273-297.
Desilets, A. R., Dhakal-Karki, S. & Dunican, K. C. 2008.
Role of metformin for weight management in patients
without type 2 diabetes. Annals of Pharmacotherapy,
42, 817-826.
Dudley, J. T., Deshpande, T. & Butte, A. J. 2011.
Exploiting drug–disease relationships for
computational drug repositioning. Briefings in
Bioinformatics, 12
, 303-311.
Feinerer, I. & Hornik, K. 2012. tm: text mining package. R
package version 0.5-7.1.
Frantz, S. 2005. Drug discovery: playing dirty. Nature,
437, 942-943.
Freund, Y. & Schapire, R. E. Experiments with a new
boosting algorithm. In: Proceedings of the 13th
International Conference on Machine Learning, 1996
Bari, Italy. 148-156.
Fürnkranz, J. 1998. A study using n-gram features for text
categorization. Austrian Research Institute for
Artifical Intelligence, 3, 1-10.
HEALTHINF 2017 - 10th International Conference on Health Informatics
116

Gadde, K. M., Parker, C. B., Maner, L. G., Wagner, H. R.,
Logue, E. J., Drezner, M. K. & Krishnan, K. R. R.
2001. Bupropion for weight loss: an investigation of
efficacy and tolerability in overweight and obese
women. Obesity Research, 9, 544-551.
Garsed, K., Chernova, J., Hastings, M., Lam, C., Marciani,
L., Singh, G., Henry, A., Hall, I., Whorwell, P. &
Spiller, R. 2014. A randomised trial of ondansetron for
the treatment of irritable bowel syndrome with
diarrhoea. Gut, 63, 1617-1625.
Gottlieb, A., Stein, G. Y., Ruppin, E. & Sharan, R. 2011.
PREDICT: a method for inferring novel drug
indications with application to personalized medicine.
Molecular Systems Biology, 7, 496.
Greenway, F. L., Fujioka, K., Plodkowski, R. A.,
Mudaliar, S., Guttadauria, M., Erickson, J., Kim, D.
D., Dunayevich, E. & Group, C.-I. S. 2010. Effect of
naltrexone plus bupropion on weight loss in
overweight and obese adults (COR-I): a multicentre,
randomised, double-blind, placebo-controlled, phase 3
trial. The Lancet, 376, 595-605.
Hall, M., Frank, E., Holmes, G., Pfahringer, B.,
Reutemann, P. & Witten, I. H. 2009. The WEKA data
mining software: an update. ACM SIGKDD
explorations newsletter, 11, 10-18.
He, H. & Garcia, E. A. 2009. Learning from imbalanced
data. IEEE Transactions on Knowledge and Data
Engineering, 21, 1263-1284.
Hu, G. & Agarwal, P. 2009. Human disease-drug network
based on genomic expression profiles. PLoS ONE, 4,
e6536.
Igel, L. I., Sinha, A., Saunders, K. H., Apovian, C. M.,
Vojta, D. & Aronne, L. J. 2016. Metformin: an old
therapy that deserves a new indication for the
treatment of obesity. Current Atherosclerosis Reports,
18, 1-8.
Jain, A. K., Kaplan, R. A., Gadde, K. M., Wadden, T. A.,
Allison, D. B., Brewer, E. R., Leadbetter, R. A.,
Richard, N., Haight, B. & Jamerson, B. D. 2002.
Bupropion SR vs. placebo for weight loss in obese
patients with depressive symptoms. Obesity Research,
10, 1049-1056.
Jensen, P. B., Jensen, L. J. & Brunak, S. 2012. Mining
electronic health records: towards better research
applications and clinical care. Nature Reviews
Genetics, 13, 395-405.
Keiser, M. J., Setola, V., Irwin, J. J., Laggner, C., Abbas,
A., Hufeisen, S. J., Jensen, N. H., Kuijer, M. B.,
Matos, R. C., Tran, T. B., Whaley, R., Glennon, R. A.,
Hert, J., Thomas, K. L. H., Edwards, D. D., Shoichet,
B. K. & Roth, B. L. 2009. Predicting new molecular
targets for known drugs. Nature, 462, 175-181.
Khatri, P., Roedder, S., Kimura, N., De Vusser, K.,
Morgan, A. A., Gong, Y., Fischbein, M. P., Robbins,
R. C., Naesens, M., Butte, A. J. & Sarwal, M. M.
2013. A common rejection module (CRM) for acute
rejection across multiple organs identifies novel
therapeutics for organ transplantation. The Journal of
Experimental Medicine, 210, 2205-2221.
Knezevic, M. Z., Bivolarevic, I. C., Peric, T. S. &
Jankovic, S. M. 2011. Using Facebook to increase
spontaneous reporting of adverse drug reactions. Drug
Safety, 34, 351-352.
Kotsiantis, S., Kanellopoulos, D. & Pintelas, P. 2006.
Handling imbalanced datasets: A review. GESTS
International Transactions on Computer Science and
Engineering, 30, 25-36.
Leaman, R., Doğan, R. I. & Lu, Z. 2013. DNorm: disease
name normalization with pairwise learning to rank.
Bioinformatics, 29, 2909-2917.
Manning, C. D., Surdeanu, M., Bauer, J., Finkel, J. R.,
Bethard, S. & Mcclosky, D. The Stanford CoreNLP
natural language processing toolkit. In: The 52nd
Annual Meeting of the Association for Computational
Linguistics: System Demonstrations, 2014 Baltimore,
MD, USA. 55-60.
Mcdonagh, M. S., Selph, S., Ozpinar, A. & Foley, C.
2014. Systematic review of the benefits and risks of
metformin in treating obesity in children aged 18 years
and younger. JAMA Pediatrics, 168, 178-184.
Michalski, R. S., Carbonell, J. G. & Mitchell, T. M. 2013.
Machine learning: An artificial intelligence approach,
Springer Science & Business Media.
Paolisso, G., Amato, L., Eccellente, R., Gambardella, A.,
Tagliamonte, M. R., Varricchio, G., Carella, C.,
Giugliano, D. & D'onofrio, F. 1998. Effect of
metformin on food intake in obese subjects. European
Journal of Clinical Investigation, 28, 441-446.
Pedersen, T., Pakhomov, S. V. S., Patwardhan, S. &
Chute, C. G. 2007. Measures of semantic similarity
and relatedness in the biomedical domain. Journal of
Biomedical Informatics, 40, 288-299.
Peirson, L., Douketis, J., Ciliska, D., Fitzpatrick-Lewis,
D., Ali, M. U. & Raina, P. 2014. Treatment for
overweight and obesity in adult populations: a
systematic review and meta-analysis. CMAJ Open, 2,
E306-E317.
Pleyer, L. & Greil, R. 2015. Digging deep into “dirty”
drugs–modulation of the methylation machinery.
Drug
Metabolism Reviews, 47, 252-279.
Powell, G. E., Seifert, H. A., Reblin, T., Burstein, P. J.,
Blowers, J., Menius, J. A., Painter, J. L., Thomas, M.,
Pierce, C. E., Rodriguez, H. W., Brownstein, J. S.,
Freifeld, C. C., Bell, H. G. & Dasgupta, N. 2016.
Social media listening for routine post-marketing
safety surveillance. Drug Safety, 39, 443-454.
Quinlan, J. R. 2014. C4.5: programs for machine learning,
Elsevier.
Ru, B., Harris, K. & Yao, L. A Content Analysis of
Patient-Reported Medication Outcomes on Social
Media. In: Proceedings of IEEE 15th International
Conference on Data Mining Workshops, 2015 Atlantic
City, NJ, USA. IEEE, 472-479.
Sánchez, D., Batet, M., Isern, D. & Valls, A. 2012.
Ontology-based semantic similarity: A new feature-
based approach. Expert Systems with Applications, 39,
7718-7728.
Sanseau, P., Agarwal, P., Barnes, M. R., Pastinen, T.,
Richards, J. B., Cardon, L. R. & Mooser, V. 2012. Use
Identifying Serendipitous Drug Usages in Patient Forum Data - A Feasibility Study
117

of genome-wide association studies for drug
repositioning. Nature Biotechnology, 30, 317-320.
Shah, N. H. & Musen, M. A. UMLS-Query: a perl module
for querying the UMLS. In: AMIA Annual Symposium,
2008 Washington, DC, USA. 652-656.
Shandrow, K. L. 2016. The Hard Truth: What Viagra Was
Really Intended For [Online]. Entrepreneur.com.
Available:
http://www.entrepreneur.com/article/254908
[Accessed 02/22/2016].
Shim, J. S. & Liu, J. O. 2014. Recent advances in drug
repositioning for the discovery of new anticancer
drugs. International Journal of Biological Sciences,
10, 654-63.
Socher, R., Perelygin, A., Wu, J. Y., Chuang, J., Manning,
C. D., Ng, A. Y. & Potts, C. Recursive deep models
for semantic compositionality over a sentiment
treebank. In: Proceedings of the Conference on
Empirical Methods in Natural Language Processing,
2013 Seattle, WA, USA. Citeseer, 1631-1642.
Stoll, A. L. & Rueter, S. 1999. Treatment augmentation
with opiates in severe and refractory major depression.
American Journal of Psychiatry, 156, 2017.
Tenore, P. L. 2008. Psychotherapeutic benefits of opioid
agonist therapy. Journal of Addictive Diseases, 27, 49-
65.
Tetsunaga, T., Tetsunaga, T., Tanaka, M. & Ozaki, T.
2015. Efficacy of tramadol–acetaminophen tablets in
low back pain patients with depression. Journal of
Orthopaedic Science, 20, 281-286.
U.S. National Library of Medicine. 2016a. MEDLINE
Fact Sheet [Online]. Available:
https://www.nlm.nih.gov/pubs/factsheets/medline.html
[Accessed 09/29/2016].
U.S. National Library of Medicine. 2016b. SNOMED CT
[Online]. Available:
http://www.nlm.nih.gov/research/umls/Snomed/snome
d_main.html [Accessed 08/03/2015].
Wren, J. D., Bekeredjian, R., Stewart, J. A., Shohet, R. V.
& Garner, H. R. 2004. Knowledge discovery by
automated identification and ranking of implicit
relationships. Bioinformatics, 20, 389-398.
Wu, T.-F., Lin, C.-J. & Weng, R. C. 2004. Probability
estimates for multi-class classification by pairwise
coupling. Journal of Machine Learning Research, 5,
975-1005.
Xu, H., Aldrich, M. C., Chen, Q., Liu, H., Peterson, N. B.,
Dai, Q., Levy, M., Shah, A., Han, X., Ruan, X., Jiang,
M., Li, Y., Julien, J. S., Warner, J., Friedman, C.,
Roden, D. M. & Denny, J. C. 2014. Validating drug
repurposing signals using electronic health records: a
case study of metformin associated with reduced
cancer mortality. Journal of the American Medical
Informatics Association, 22, 179-191.
Yang, C. C., Yang, H., Jiang, L. & Zhang, M. Social
media mining for drug safety signal detection. In:
Proceedings of the 2012 International Workshop on
Smart Health and Wellbeing, 2012 Maui, HI, USA.
ACM, 33-40.
Yao, L. & Rzhetsky, A. 2008. Quantitative systems-level
determinants of human genes targeted by successful
drugs. Genome Research, 18, 206-213.
Yao, L., Zhang, Y., Li, Y., Sanseau, P. & Agarwal, P.
2011. Electronic health records: Implications for drug
discovery. Drug Discovery Today, 16, 594-599.
Yates, A. & Goharian, N. ADRTrace: detecting expected
and unexpected adverse drug reactions from user
reviews on social media sites. In: The 35th European
Conference on Information Retrieval, 2013 Moscow,
Russia. Springer-Verlag, 816-819.
HEALTHINF 2017 - 10th International Conference on Health Informatics
118
