
Thrombus Detection in CT Brain Scans using a Convolutional Neural
Network
Aneta Lisowska
1,2
, Erin Beveridge
1
, Keith Muir
3
and Ian Poole
1
1
Toshiba Medical Visualization Systems Europe Ltd., 2 Anderson Place, Edinburgh, U.K.
2
School of Engineering and Physical Sciences, Heriot-Watt University, Edinburgh, U.K.
3
Queen Elizabeth University Hospital, University of Glasgow, Glasgow, U.K.
Keywords:
Convolutional Neural Network, Stroke, Thrombus, Non-contrast Computer Tomography (NCCT).
Abstract:
Automatic detection and measurement of thrombi may expedite clinical workflow in the treatment planning
stage. Nevertheless it is a challenging task on non-contrast computed tomography due to the subtlety of
the pathological intensity changes, which are further confounded by the appearance of vascular calcification
(common in ageing brains). In this paper we propose a 3D Convolutional Neural Network architecture to detect
these subtle signs of stroke. The architecture is designed to exploit contralateral features and anatomical atlas
information. We use 122 CT volumes equally split into training and testing to validate our method, achieving
a ROC AUC of 0.996 and a Precision-Recall AUC of 0.563 in a voxel-level evaluation. The results are not yet
at a level for routine clinical use, but they are encouraging.
1 INTRODUCTION
Detection of subtly discriminated regions in volumet-
ric medical imaging is challenging. An example is
the detection of stroke signs in non-contrast computed
tomography (NCCT), where thrombi are one of the
pathologies of interest. Thrombi manifest as subtle
vascular intensity and texture changes which are chal-
lenging to identify due to the proximity of bone, and
the similarity to normal age related vascular calcifica-
tion (see Figure 1).
Automatic detection of thrombi and their subse-
quent measurements may provide information which
is important for treatment planning. For example, the
length of the thrombus observed in thin slice NCCT is
related to the success rate of thrombolysis, a treatment
aimed to recanalise occluded arteries (Riedel et al.,
2011). Nevertheless, to our knowledge, the problem
of automatic thrombus detection has not been directly
addressed.
Detection of stroke signs in CT scans has previ-
ously been demonstrated at the brain level. Chawla
et al. proposed a general system for classification
of ischaemic and haemorrhagic stroke signs (Chawla
et al., 2009). Their system computes a separate his-
togram of intensity values for each hemisphere, and
the classification of the image slice is based on the
comparison between the left and right histograms.
The authors report good performance in determining
the type of stroke, but the system is not designed to
Figure 1: Left: Normal calcification of arteries, which could
be confused with abnormal dense vessel signs. Right: Ex-
ample of a CT brain scan with a clearly visible dense vessel.
give a precise location of the stoke sign. Furthermore
the haemorrhage is easily detectable on CT compared
to ischaemia or thrombus.
Precise location of stroke signs requires a more lo-
calised model. One approach is to compare the patient
image with a normative atlas created from healthy ex-
amples of anatomy; the patient image is registered to
the normative atlas and the pathology is identified by
the differences between the reference atlas and the ex-
amined image (Doyle et al., 2013). This approach re-
quires accurate non-rigid registration, which may not
always be possible due to anatomical variability. Ma-
chine learning algorithms are one method of learning
diverse non-parametric appearance models. For in-
stance, random forest classifiers have been success-
fully applied to various medical imaging segmenta-
tion tasks (Geremia et al., 2011; Zikic et al., 2012).
However, in recent years CNN-based solutions have
dominated brain image analysis applications, result-
ing in the best performance in the Ischemic Stroke Le-
24
Lisowska A., Beveridge E., Muir K. and Poole I.
Thrombus Detection in CT Brain Scans using a Convolutional Neural Network.
DOI: 10.5220/0006114600240033
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 24-33
ISBN: 978-989-758-215-8
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

sion Segmentation (ISLES) (winning solution in 2015
(Kamnitsas et al., 2016)) and Brain Tumor Segmenta-
tion (BRATS) challenges (see CNN based application
(Havaei et al., 2016) (Pereira et al., 2015)). These
CNN solutions are trained on brain MRI data, how-
ever CNNs have also been applied to CT images of the
knee (Prasoon et al., 2013) and lungs (Anthimopoulos
et al., 2016). The CNN architecture design may differ
between the imaging modalities and between various
anatomies of interest. (Li et al., 2014) observe that CT
images lack structural detail in regions of soft tissue,
taking on a textured appearance, therefore there are
few higher-level concepts for a deep network to learn.
Prasoon et al. also avoided use of deep architecture
in CT and pointed out that thin structure of cartilage
can be lost if too many mean-pooling layers are ap-
plied (Prasoon et al., 2013). In this paper we employ
a fully convolutional architecture, which is only 7 lay-
ers deep as described in Section 2.
CT scans are three-dimensional (3D), and there-
fore architectures designed for images cannot be di-
rectly applied. When moving to 3D data, three strate-
gies are common. Firstly, the volume may be treated
as a stack of 2D slices on which a 2D architecture
is directly applied. Volumes are often acquired in
a slice-wise manner, and where the slice spacing is
large, this may be a natural approach. The second
strategy is a triplanar CNN (Prasoon et al., 2013;
Yang et al., 2015) in which axial, sagittal and coro-
nal slices centred at the voxel of interest are consid-
ered separately and fed to three 2D CNNs (or three
channels of a 2D CNN (Roth et al., 2014; Wolterink
et al., 2015)). Their outputs are combined in the pre-
classification stage. Finally, 3D convolution (Urban
et al., 2014; Kamnitsas et al., 2015; Payan and Mon-
tana, 2015) may be used in place of 2D convolution.
This last strategy is computationally expensive, thus
training and optimisation of a network can be ardu-
ous. Payan and Montana compared a 2D CNN with
a 3D CNN on brain MRI data and found that the 3D
CNN obtained marginally better Alzheimer classifica-
tion accuracy (Payan and Montana, 2015). In this pa-
per we adopt a 3D CNN, but we apply spatial decom-
position of the kernels to address the computational
complexity issue such that convolutions are applied
one dimension at the time.
A key element of our architecture is bilateral fea-
ture comparison. The idea of bilateral feature compar-
ison is strongly linked with exploitation of anatom-
ical symmetry, which has been also incorporated in
unsupervised approaches of pathology detection. Re-
searchers utilised in-organ symmetry for detection of
tumours in the brain (Hasan et al., 2016; Zhao et al.,
2013) and prostate (Qiu et al., 2013). There have also
been attempts to examine symmetry between a pair of
organs in breast tumour detection (Erihov et al., 2015;
Bandyopadhyay, 2010). In these approaches, the
pathology is found by searching for the most dissimi-
lar regions between the left and right side of the organ
(Hasan et al., 2016), or by identification of asymmetry
between paired organs (Erihov et al., 2015). It is rare
that a stroke will occur in both sides of the brain in
a given episode, therefore comparison between hemi-
spheres helps to discriminate between subtle changes
in the affected side and the normal brain tissue. We
hypothesise that exploiting contralateral features will
therefore be useful when detecting thrombi.
The second key element of our architecture is the
explicit incorporation of spatial anatomical context by
provision of the coordinates in a (rigidly-registered)
reference dataset “atlas” by adding three channels
encoding x, y, z atlas locations. In this, we fol-
low the lead of authors such as (O’Neil et al., 2015)
who used atlas coordinates as a feature provided to
a random forest when learning to detect anatomi-
cal landmarks. The injection of contextual informa-
tion into a machine learning algorithm was demon-
strated in a traumatic brain injury application by (Rao
et al., 2014) who used multi-atlas propagation and
EM (MALPEM) (Ledig et al., 2012) to obtain tissue
classes priors which were later exploited in a random
forest. Since the thrombi may be subtle, the addition
of anatomical context might provide a helpful insight.
One of the challenges in training a CNN to de-
tect thrombus is the significantly limited set of sam-
ples containing pathology compared to healthy tis-
sue. As a result, the CNN may take little account of
the underrepresented class during training. One solu-
tion to address this problem is to alter the loss func-
tion. For neutrophil detection, (Wang et al., 2015)
penalised more heavily the misclassification of non-
neutrophils than that of neutrophils, by doubling the
computed softmax loss. (Brosch et al., 2015) de-
signed a specialised CNN objective function which
relies on a weighted combination of sensitivity and
specificity. A more mechanical approach is to take
all samples from the underrepresented category and a
random subset of the examples from the other cate-
gory (Ciresan et al., 2012; Roth et al., 2014; Li et al.,
2015; Rao et al., 2015). However, random sampling
of classes may not always give the most useful train-
ing distribution. A number of papers deliberately bi-
ased the training distribution towards examples which
were more difficult to classify due to visual proxim-
ity between classes. Researchers commonly sampled
background more densely close to the region of in-
terest compared to other areas (Ciresan et al., 2013;
Prasoon et al., 2013; Dvorak and Menze, 2015). Re-
Thrombus Detection in CT Brain Scans using a Convolutional Neural Network
25

cently, (Kamnitsas et al., 2016) suggested that train-
ing on large image segments allows for automatic ad-
justment to this problem. In this paper we choose to
use a weighting function to compensate for the unbal-
anced sample distribution such that normal and abnor-
mal samples count equally in the loss function.
In summary, this paper makes three contributions:
• We propose a 3D CNN architecture incorporating
brain symmetry exploitation and atlas information
( Section 2). We present qualitative and quanti-
tative results obtained by applying the proposed
CNN to the thrombus detection problem ( Section
4.2).
• As far as we are aware, this is the first CNN-based
solution applied to brain NCCT and the first solu-
tion for automatic thrombus detection.
• We study the effect of the training procedure on
efficiency. We compare patch and segment based
training to full block training with and without
weighting of the loss function in order to identify
the most effective training strategy in an unbal-
anced sample setting ( Section 4.1).
2 METHODS
We propose a convolutional neural network (CNN)
which leverages left and right hemisphere compari-
son (See Figure 3). The first steps consist of isotropic
resampling to 1mm voxels and alignment of the CT
volumes to a reference dataset designated as an atlas.
The transformation between a given dataset and a ref-
erence atlas is discovered via landmarks, which are
detected in the novel volume by random forest as pro-
posed in (Dabbah et al., 2014). A block of interest
extending into the sagittal plane is then extracted and
folded along the brain midline, similarly to the fold-
ing of a butterfly’s wings (see Figure 2). This folding
results in two 3D CNN intensity channels relating to
the target and contralateral sides of the brain, enabling
bilateral comparison. We call this architecture a But-
terfly CNN. Since the architecture works in the folded
space, it is left-right agnostic with respect to the tar-
get laterality. The target and contralateral channels
may thus correspond to either left and right, or right
and left, respectively. Consequently there is sharing
of training data between left and right hemispheres.
Alongside the two data input channels, we insert
three channels encoding the x, y and z atlas coordi-
nates to the architecture. For the convolution opera-
tions, we use a spatial decomposition of a 5x5x5 fil-
ter e.g. three layers of orthogonal one-dimensional
convolutions: 5x1x1, 1x5x1, 1x1x5. N
I
= 32 ker-
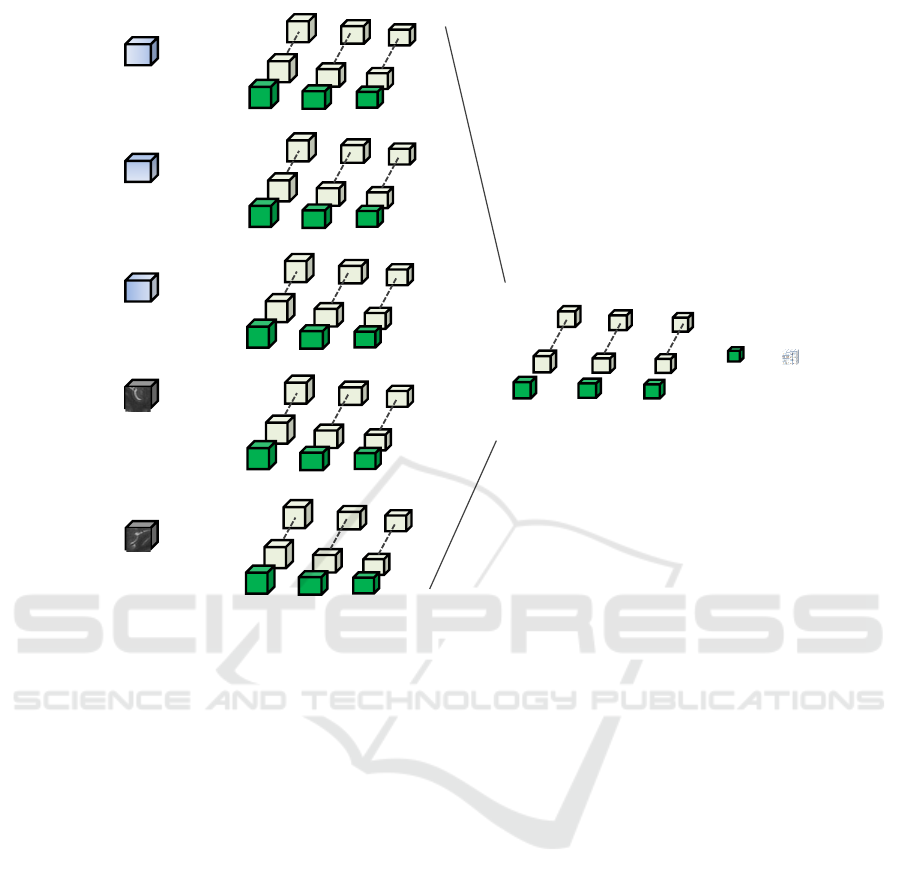
Figure 2: Block extraction.
nels are used for the data channels and N
A
= 4 ker-
nels are used for the atlas channels. Channels are then
merged and another convolution operation is applied,
with N
M
= 32 kernels. See Figure 3 for a diagram of
the architecture. ReLU activation functions are used.
The output is fully convolutional allowing for the effi-
cient prediction of all voxels of the dataset in a single
pass.
The model was implemented in Python using the
Keras library built on top of Theano. Training was
performed using stochastic gradient descent on nor-
malised data samples, with learning rate 0.001, op-
timising the squared hinge loss function, momentum
0.9 and L2 regularisation 0.002.
3 MATERIALS
We use data from the South Glasgow Stroke Imag-
ing Database, which includes the following studies:
ATTEST (Huang et al., 2015), POSH and WYETH
(Wardlaw et al., 2013) provided by the Institute of
Neuroscience and Psychology, University of Glas-
gow, Southern General Hospital. Ground truth was
collected on the acute non-contrast CT (NCCT) scan
for 122 patients with suspected acute ischaemic stroke
within 6 hours of onset. Manual segmentations of
thrombi were generated in 3D Slicer 4.5.0 by a clin-
ical researcher under the supervision of an experi-
enced neuroradiologist. Annotations were blind to
additional scans (e.g. CT angiography, CT perfusion,
follow-up scans) and clinical information with the ex-
ception of the laterality of the symptoms. Current
methods are applied only to the anterior circulation
but consider both proximal occlusions and more dis-
tal dot signs.
There might be some uncertainty around precise
segmentation of the thrombi. Since we are interested
only in detection, it is undesirable for the network to
waste learning capacity trying to refine the boundary.
We consider a border, 2mm in width around each ab-
normality, as a “do not care” zone. Such voxels do not
influence the loss function which drives the stochastic
gradient descent, nor are they included in the results.
BIOIMAGING 2017 - 4th International Conference on Bioimaging
26
abs(x)
y
z
CNN Input
1 @ 1 x 1 x 1
CNN Output
𝑁
𝑀
@ 5 x 1 x 1
𝑁
𝑀
@ 1 x 5 x 1 𝑁
𝑀
@ 1 x 1 x 5
Target
laterality
Comparative
laterality
Prediction
for target
laterality
𝑁
𝐼
@ 5 x 1 x 1
𝑁
𝐼
@ 1 x 5 x 1 𝑁
𝐼
@ 1 x 1 x 5
𝑁
𝐴
@ 5 x 1 x 1 𝑁
𝐴
@ 1 x 5x 1 𝑁
𝐴
@ 1 x 1 x 5
𝑁
𝐴
@ 5 x 1 x 1
𝑁
𝐴
@ 1 x 5x 1
𝑁
𝐴
@ 1 x 1 x 5
𝑁
𝐴
@ 5 x 1 x 1 𝑁
𝐴
@ 1 x 5x 1
𝑁
𝐴
@ 1 x 1 x 5
𝑁
𝐼
@ 5 x 1 x 1
𝑁
𝐼
@ 1 x 5 x 1 𝑁
𝐼
@ 1 x 1 x 5
Figure 3: Schematic of the CNN architecture.
For implementation, we use -1 and +1 to represent
normal and thrombi voxels respectively, and use the
value 0 to represent the ”do not care” voxels. The
border zone is constructed using morphological dila-
tion.
For training and testing of the CNN classifier we
used a Titan X GPU.
4 EXPERIMENTS
The data was split equally (and randomly) into 61
training datasets and 61 testing datasets.
We evaluated the performance of the CNN in Sec-
tion 4.2 in terms of the area under the curve (AUC)
for the Receiver Operating Curve (ROC AUC) and
the precision-recall (PR AUC) curve. The ROC curve
represents the relationship between sensitivity (i.e. re-
call) and specificity, while the PR curve represents the
relationship between precision and recall. Sensitiv-
ity (recall) is a measure of how many of the positive
samples have been correctly identified as being pos-
itive. Specificity is a measure of how many of the
negative samples have been correctly identified as be-
ing negative. Precision is a measure of how many of
the samples predicted by the classifier as positive are
indeed positive. It is suggested that PR curves should
be used when the positive class samples are rare com-
pared to the negative class samples (Davis and Goad-
rich, 2006), because precision is more sensitive to any
change in the number of false positives, while speci-
ficity is not due to the large number of negative sam-
ples. As an example, if the ratio of normal to abnor-
mal is 10, 000 : 1 then even a false positive rate of
0.01 results in 100 false detections.
4.1 Patch vs. Full Block Training
In our efforts to establish the most efficient train-
ing procedure, we compare training of the CNN with
whole blocks as input versus training with small
patches as input. We introduce a weighting factor w to
allow for the correction of the imbalance between nor-
mal and abnormal samples. We define w as the ratio
of normal to abnormal voxels in the training set. The
CNN architecture as presented in Figure 3 is used, but
with smaller filter size 3x3x3 and trained only for 250
epochs. We are primarily interested in the training
time of the network, but we also report PR AUC to
ensure the learning took place.
Thrombus Detection in CT Brain Scans using a Convolutional Neural Network
27
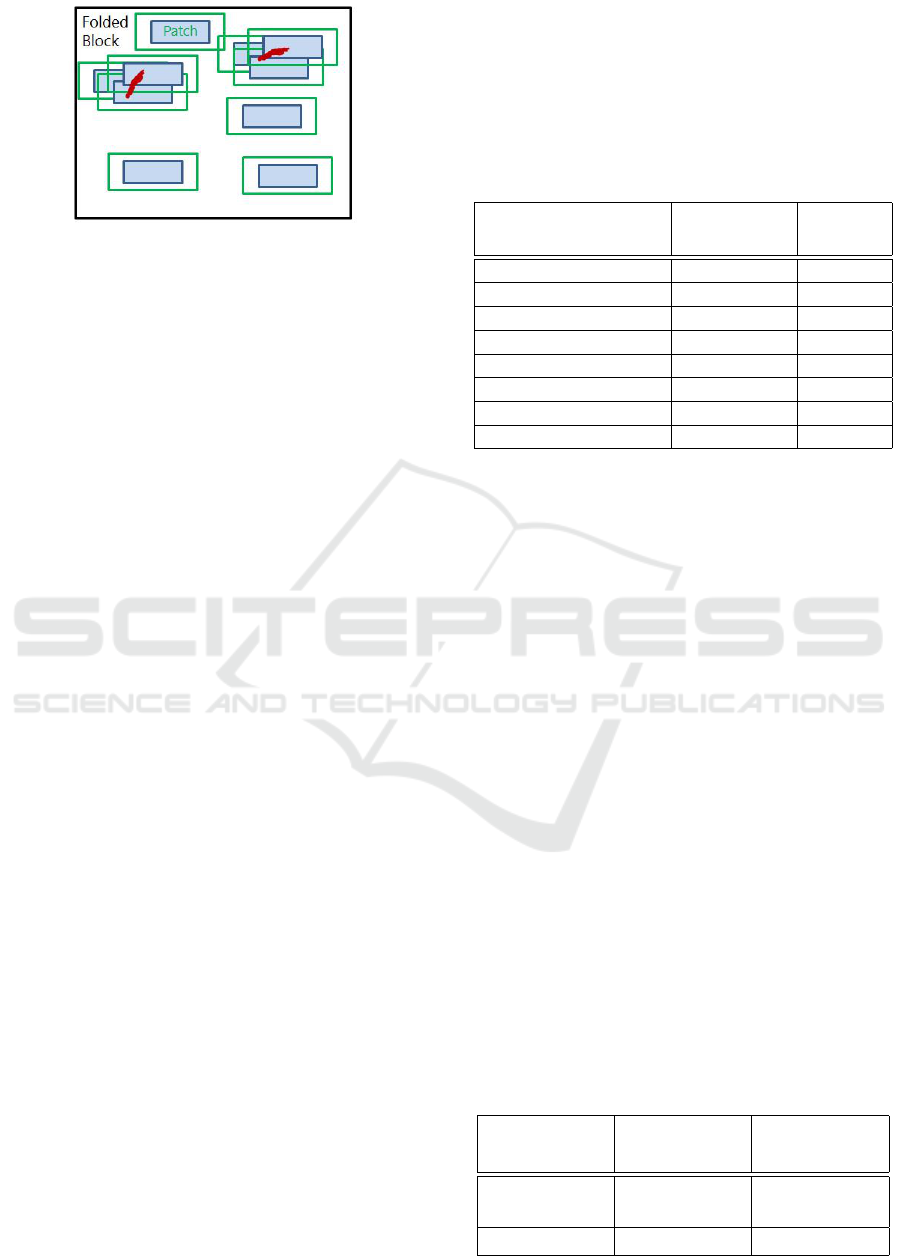
Figure 4: 2D representation of biased patch selection pro-
cess. More patches of abnormality than normality are se-
lected, in order to compensate for the presence of normal
voxels within the abnormal patches, to achieve a balanced
training set. The green box represents the field of view (a
margin) and the blue box is the core patch size, for which
predictions are being output during training.
For the patch training, we sample equal numbers
of normal and abnormal locations, but we vary the
size of the core patch that we extract at each location.
The core size corresponds to the voxels for which the
response is computed. In practice, we require a larger
input patch to the network, in order to include a mar-
gin of voxels for which the filter response is not com-
puted, as we employ valid-mode convolutional oper-
ations. In one dimension and input patch size P, con-
volving with filter F will produce a feature map M of
size P − F + 1. Therefore, to produce an output of
1 voxel after 2 convolutions with filter size 3 in each
dimension, the input patch size is 5
3
. The core patch
size of 1 will have samples only from one class, never-
theless inputs with a larger core patch size are likely
to have voxels from both classes when an abnormal
patch is extracted (See Figure 4. This case resembles
the image segment training procedure of (Kamnitsas
et al., 2016). The results obtained for 3 different in-
put patch sizes and full folded block training are pre-
sented in Table 1.
We found that full block training took significantly
longer than patch-based training. It is evident that
weighting of the loss function is necessary to compen-
sate any severe imbalance of training data samples, as
is the case when the CNN is trained on the full block
or large patches. For the core patch size 1
3
, no learn-
ing occurred when using either a weighted or an un-
weighted loss function. It might be due to exposure
of the network to insufficient number of training vox-
els, which is not a problem when trained on the larger
patches. The network trained on core patch size 1
3
might not have seen the difficult examples which are
close to the lesion borders, which would always be
included when training on the larger patches.
In a further experiment we train the CNN on large
patches with a weighted loss function. This allows for
efficient training without the need for extensive ex-
perimentation to select an appropriate patch size, for
which weighting of the loss function is not required.
Table 1: The table shows the precision-recall AUC achieved
on a test dataset after just 250 epochs of training and cor-
responding training time. PBT (Patch based training), FBT
(Full Block Training), PBTw (Patch based training with loss
function weighting), FBTw (Full Block Training with loss
function weighting).
Method Precision-
Recall AUC
Training
time
PBT (core patch 1) 0.015 1.50 h
PBTw (core patch 1) 0.023 1.52 h
PBT (core patch 5) 0.341 29.3 min
PBTw(core patch 5) 0.320 29.4 min
PBT (core patch 10) 0.006 36.4 min
PBTw (core patch 10) 0.347 36.2 min
FBT 0.002 16.7 h
FBTw 0.397 16.6 h
4.2 Thrombus Detection Results
The CNN detector (as presented in Figure 3) was
trained for 800 epochs on input patches of size 24
3
(core patch 16
3
) extracted from 61 training datasets.
The detector was evaluated on 61 testing datasets
and the quantitative results are presented in Table 2.
Training took 2.15h and detection time/ per block is
4s.
For comparison we trained also a simple CNN net-
work which is composed from 7 layers and the num-
ber of kernels and filter sizes are the same as in the
Figure 3 the difference is that it has only one input
channel. The bilateral comparison and atlas informa-
tion is not incorporated in the simple CNN.
The precision-recall curves for both the simple
and the Butterfly CNNs are shown in Figure 5 and
the ROC for the Butterfly CNN is shown in Figure
6. Some example detection results are presented in
Figure 7 and Figure 8. The former show a case of
very subtle thrombus where we fail to detect it at the
sagittal slice of the abnormality and quite a few false
positives are visible in the maximum intensity pro-
jection (MIP) view. The latter is more clearly visible
thrombus and the detections are almost perfect.
Table 2: Thrombus detection evaluation.
Model ROC AUC Precision-
Recall AUC
Butterfly
CNN
0.997 0.562
Simple CNN 0.996 0.178
BIOIMAGING 2017 - 4th International Conference on Bioimaging
28
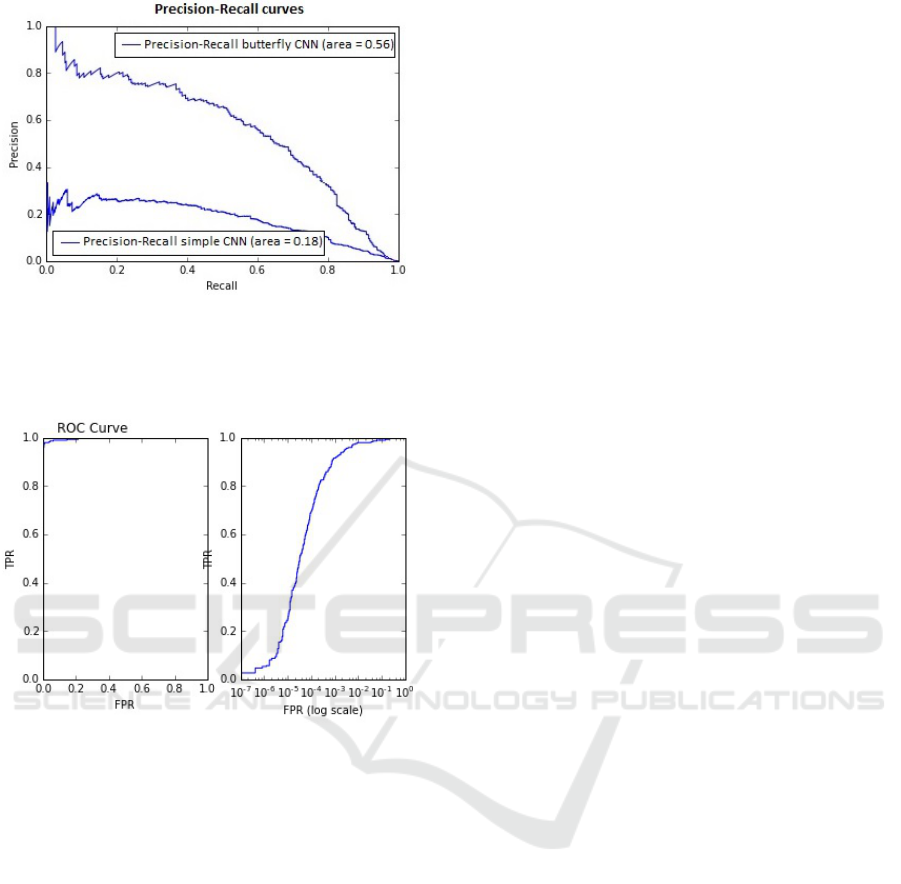
Figure 5: Precision-Recall curves for Simple CNN and But-
terfly CNN. At the point that around 70% of thrombi are
detected the simple CNN would give around 20% true de-
tection results whereas the Butterfly CNN would produce
just above 50% of true detection results.
Figure 6: Left: ROC curve in full. Right: ROC curve shown
with a log scale on the specificity axis, to focus on the range
of interest. For the thrombus detector to be clinically useful
the false positive rate at the voxel level needs to be very low,
of order 10
−4
or better, otherwise false positive detections
will overwhelm true positives.
5 DISCUSSION
We have found that weighting of the loss function to
compensate for unbalanced training data is effective,
even essential, when training either on the full block
or using a large image patches. Patch-based training
is significantly faster than full block training. The
downside of this approach is that it requires careful
selection of the size of the patches when the weighting
of the loss function is not applied. To overcome this
difficulty we have adapted the combination of larger
path and weighting of the loss function for training of
the thrombus detector.
The quantitative and visual results obtained us-
ing presented solution are encouraging. However, to
reach detection performance level at which the detec-
tor could be applied clinically the number of the false
positive detections needs to be further reduced. To
achieve this we will consider adapting a cascade ap-
proach similar to boosting, so that a second classifier
is trained only on on normal examples which were
misclassified at the first detection stage, this might
help to eliminate false positive detections (Liswoska
et al., 2016).
The proposed architecture for thrombus detection
performs better than a single channel CNN. This sup-
ports our hypothesis that incorporation of atlas in-
formation and bilateral features within the CNN ar-
chitecture is helpful when identifying subtle stroke
signs. In future experiments it would be interesting to
evaluate the extent to which each of the architectural
choices contribute to the thrombus detection perfor-
mance and whether the level at which they are in-
serted to the CNN architecture affects their useful-
ness.
Large amount of medical imaging data is diffi-
cult to obtain, especially given the requirement for
ground truth labels provided by expert clinicians. Re-
searchers have tried to address this problem by us-
ing transfer learning. In this scheme a network first
trained on large annotated natural image datasets like
ImageNet, followed by fine-tuning on the smaller
medical imaging dataset. (Shin et al., 2016) found this
approach to be beneficial when applied to both tho-
racoabdominal lymph node detection and interstitial
lung disease classification in CT scans. On the other
hand (Arevalo et al., 2015) and (Gao et al., 2016)
found that CNN models trained fully with the domain
images performed better than models pretrained on
the ImageNet dataset when applied to the problems
of mammography mass lesion classification and Inter-
stitial Lung Disease Pattern Detection in CT respec-
tively. It is unclear if this training approach would be
helpful for thrombus detection; further investigation
is required. An interesting avenue to explore might
be knowledge transfer between two medical image
datasets or alternatively between two different tasks.
We hypothesise that the proposed CNN architec-
ture would also be well suited to the detection of other
stoke signs and we plan to train it on ischaemic re-
gions in future study. This will provide an opportu-
nity to evaluate benefits coming from within-domain
transfer learning. The model trained purely on is-
chaemic data can be compared to the thrombus detec-
tion model fine-tuned for ischaemia detection prob-
lems.
Future assessment of the detector performance
will also include patient level evaluation and testing
on more brain CT datasets from different institutions.
Thrombus Detection in CT Brain Scans using a Convolutional Neural Network
29

Figure 7: Top to bottom: CT image at the abnormal slice, ground truth segmentations at the abnormal slice (here: gray region
is a ”do not care” zone), detection at the abnormal slice (note misdetection in a sagittal slice), ground truth in a projection
view through the whole volume, detection in a projection through the volume (a few false positive detections are present).
BIOIMAGING 2017 - 4th International Conference on Bioimaging
30
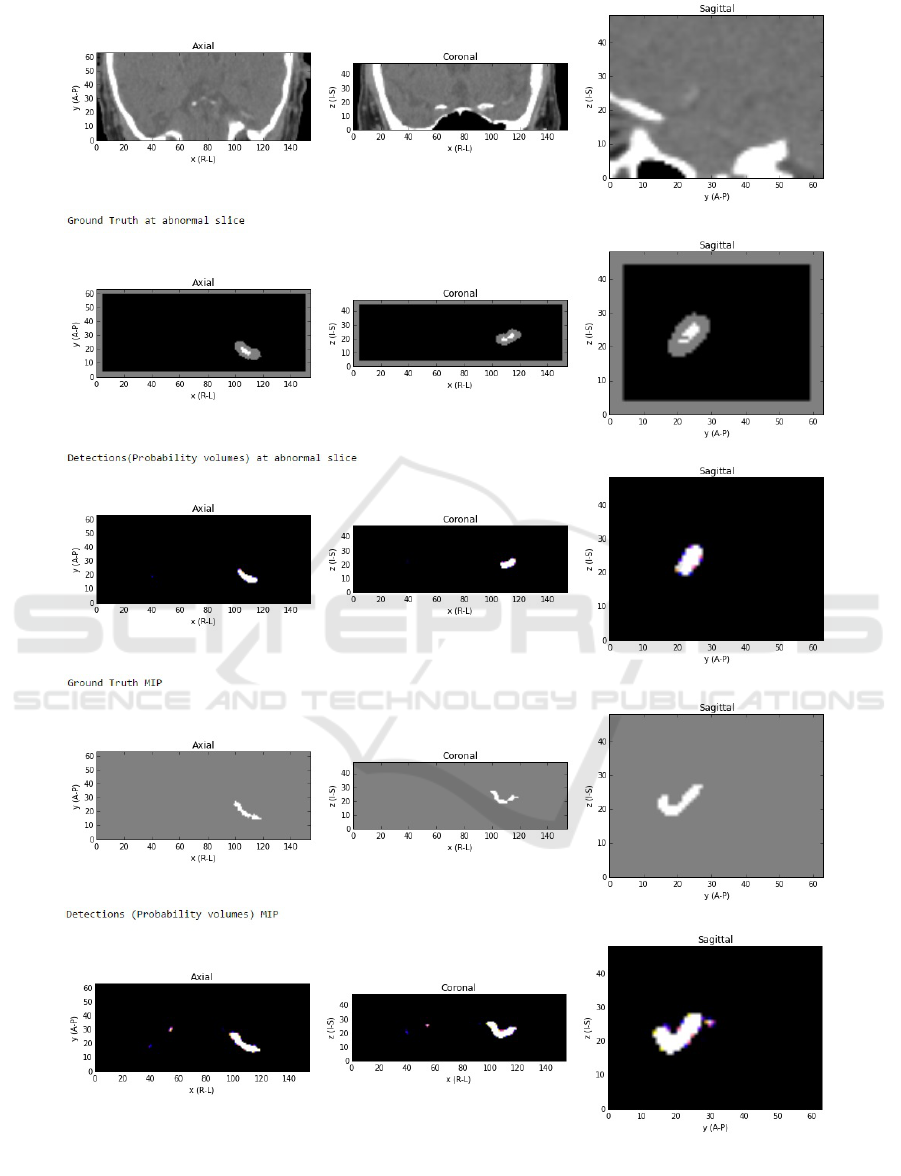
Figure 8: Top to bottom: CT image at the abnormal slice (** note posterior top, anterior bottom of the CT image) with
thrombus clearly visible, ground truth segmentations at the abnormal slice (here:gray region is a ”do not care” zone), detection
at the abnormal slice (clear detection), ground truth in a projection view through the whole volume, detection in a projection
through the volume (a few false positive detections are present, but with lower certainty (dark blue and yellow colors as
opposed to bright white true positive detections).
Thrombus Detection in CT Brain Scans using a Convolutional Neural Network
31

6 CONCLUSION
We have proposed a CNN for thrombus detection
in brain CT. The suggested architecture utilises con-
tralateral features and atlas information. The method
is evaluated at the voxel level on 61 NCCT datasets
and some example detection results are presented.
Both quantitative and visual results are encouraging.
Further investigation, as mentioned in the discussion,
may lead to improvement of the detector to a level at
which it could be applied in the clinical setting.
ACKNOWLEDGEMENTS
We wish to thank Alison O’Neil for her helpful com-
ments on the manuscript draft.
REFERENCES
Anthimopoulos, M., Christodoulidis, S., Ebner, L., Christe,
A., and Mougiakakou, S. (2016). Lung pattern classi-
fication for interstitial lung diseases using a deep con-
volutional neural network. IEEE transactions on med-
ical imaging, 35(5):1207–1216.
Arevalo, J., Gonzalez, F. A., Ramos-Pollan, R., Oliveira,
J. L., and Guevara Lopez, M. A. (2015). Convolu-
tional neural networks for mammography mass lesion
classification. In Engineering in Medicine and Biol-
ogy Society (EMBC), 2015 37th Annual International
Conference of the IEEE, pages 797–800. IEEE.
Bandyopadhyay, S. K. (2010). Breast asymmetry-tutorial
review. Breast, 9(8).
Brosch, T., Yoo, Y., Tang, L. Y., Li, D. K., Traboulsee,
A., and Tam, R. (2015). Deep convolutional encoder
networks for multiple sclerosis lesion segmentation.
In Medical Image Computing and Computer-Assisted
Intervention–MICCAI 2015, pages 3–11. Springer.
Chawla, M., Sharma, S., Sivaswamy, J., and Kishore, L.
(2009). A method for automatic detection and classi-
fication of stroke from brain ct images. In Engineering
in Medicine and Biology Society, volume 2009, pages
3581–3584.
Ciresan, D., Giusti, A., Gambardella, L. M., and Schmid-
huber, J. (2012). Deep neural networks segment neu-
ronal membranes in electron microscopy images. In
Advances in neural information processing systems,
pages 2843–2851.
Ciresan, D. C., Giusti, A., Gambardella, L. M., and Schmid-
huber, J. (2013). Mitosis detection in breast cancer
histology images with deep neural networks. Lecture
Notes in Computer Science (including subseries Lec-
ture Notes in Artificial Intelligence and Lecture Notes
in Bioinformatics), 8150 LNCS(PART 2):411–418.
Dabbah, M. A., Murphy, S., Pello, H., Courbon, R., Bev-
eridge, E., Wiseman, S., Wyeth, D., and Poole, I.
(2014). Detection and location of 127 anatomical
landmarks in diverse ct datasets. In SPIE Medical
Imaging, pages 903415–903415. International Society
for Optics and Photonics.
Davis, J. and Goadrich, M. (2006). The relationship be-
tween precision-recall and roc curves. In Proceed-
ings of the 23rd International Conference on Machine
Learning, ICML’06, pages 233–240.
Doyle, S., Vasseur, F., Dojat, M., and Forbes, F. (2013).
Fully automatic brain tumor segmentation from multi-
ple mr sequences using hidden markov fields and vari-
ational em. Procs. NCI-MICCAI BraTS, pages 18–22.
Dvorak, P. and Menze, B. (2015). Structured prediction
with convolutional neural networks for multimodal
brain tumor segmentation. Proceedings of BRATS-
MICCAI.
Erihov, M., Alpert, S., Kisilev, P., and Hashoul, S. (2015).
A cross saliency approach to asymmetry-based tumor
detection. In International Conference on Medical Im-
age Computing and Computer-Assisted Intervention,
pages 636–643. Springer.
Gao, M., Xu, Z., Lu, L., Harrison, A. P., Summers, R. M.,
and Mollura, D. J. (2016). Multi-label deep regression
and unordered pooling for holistic interstitial lung dis-
ease pattern detection. In International Workshop on
Machine Learning in Medical Imaging, pages 147–
155. Springer.
Geremia, E., Clatz, O., Menze, B. H., Konukoglu, E., Cri-
minisi, A., and Ayache, N. (2011). Spatial decision
forests for ms lesion segmentation in multi-channel
magnetic resonance images. NeuroImage, 57(2):378–
390.
Hasan, A., Meziane, F., and Khadim, M. (2016). Auto-
mated segmentation of tumours in mri brain scans. In
Proceedings of the 9th International Joint Conference
on Biomedical Engineering Systems and Technologies
(BIOSTEC 2016), pages 55–62. SCITEPRESS.
Havaei, M., Davy, A., Warde-Farley, D., Biard, A.,
Courville, A., Bengio, Y., Pal, C., Jodoin, P.-M., and
Larochelle, H. (2016). Brain tumor segmentation with
deep neural networks. Medical Image Analysis.
Huang, X., Cheripelli, B. K., Lloyd, S. M., Kalladka, D.,
Moreton, F. C., Siddiqui, A., Ford, I., and Muir, K. W.
(2015). Alteplase versus tenecteplase for thrombol-
ysis after ischaemic stroke (attest): a phase 2, ran-
domised, open-label, blinded endpoint study. The
Lancet Neurology, 14(4):368–376.
Kamnitsas, K., Chen, L., Ledig, C., Rueckert, D., and
Glocker, B. (2015). Multi-scale 3D convolutional neu-
ral networks for lesion segmentation in brain MRI. Is-
chemic Stroke Lesion Segmentation, page 13.
Kamnitsas, K., Ledig, C., Newcombe, V. F., Simpson,
J. P., Kane, A. D., Menon, D. K., Rueckert, D., and
Glocker, B. (2016). Efficient multi-scale 3d cnn with
fully connected crf for accurate brain lesion segmen-
tation. arXiv preprint arXiv:1603.05959.
Ledig, C., Wolz, R., Aljabar, P., L
¨
otj
¨
onen, J., Hecke-
mann, R. A., Hammers, A., and Rueckert, D. (2012).
Multi-class brain segmentation using atlas propaga-
tion and em-based refinement. In 2012 9th IEEE Inter-
BIOIMAGING 2017 - 4th International Conference on Bioimaging
32

national Symposium on Biomedical Imaging (ISBI),
pages 896–899. IEEE.
Li, Q., Cai, W., Wang, X., Zhou, Y., Feng, D. D., and
Chen, M. (2014). Medical image classification with
convolutional neural network. In Control Automation
Robotics & Vision (ICARCV), 2014 13th International
Conference on, pages 844–848. IEEE.
Li, W., Jia, F., and Hu, Q. (2015). Automatic segmentation
of liver tumor in CT images with deep convolutional
neural networks. Journal of Computer and Communi-
cations, 3(11):146.
Liswoska, A., Beveridge, E., and Poole, I. (2016). False
positive reduction methods applied to dense vessel de-
tection in brain ct images. Poster presented at Medical
Imaging Summer School, Favigana, Italy.
O’Neil, A., Murphy, S., and Poole, I. (2015). Anatomical
landmark detection in ct data by learned atlas location
autocontext. In MIUA, pages 189–194.
Payan, A. and Montana, G. (2015). Predicting alzheimer’s
disease: a neuroimaging study with 3d convolutional
neural networks. arXiv preprint arXiv:1502.02506.
Pereira, S., Pinto, A., Alves, V., and Silva, C. A. (2015).
Deep convolutional neural networks for the segmen-
tation of gliomas in multi-sequence mri. In Inter-
national Workshop on Brainlesion: Glioma, Multiple
Sclerosis, Stroke and Traumatic Brain Injuries, pages
131–143. Springer.
Prasoon, A., Petersen, K., Igel, C., Lauze, F., Dam, E.,
and Nielsen, M. (2013). Deep feature learning for
knee cartilage segmentation using a triplanar convolu-
tional neural network. In Medical Image Computing
and Computer-Assisted Intervention–MICCAI 2013,
pages 246–253. Springer.
Qiu, W., Yuan, J., Ukwatta, E., Sun, Y., Rajchl, M., and Fen-
ster, A. (2013). Fast globally optimal segmentation
of 3d prostate mri with axial symmetry prior. In In-
ternational Conference on Medical Image Computing
and Computer-Assisted Intervention, pages 198–205.
Springer.
Rao, A., Ledig, C., Newcombe, V., Menon, D., and Rueck-
ert, D. (2014). Contusion segmentation from subjects
with traumatic brain injury: a random forest frame-
work. In 2014 IEEE 11th International Symposium
on Biomedical Imaging (ISBI), pages 333–336. IEEE.
Rao, V., Sarabi, M. S., and Jaiswal, A. (2015). Brain tu-
mor segmentation with deep learning. Proceedings of
BRATS-MICCAI.
Riedel, C. H., Zimmermann, P., Jensen-Kondering, U., Stin-
gele, R., Deuschl, G., and Jansen, O. (2011). The
importance of size successful recanalization by intra-
venous thrombolysis in acute anterior stroke depends
on thrombus length. Stroke, 42(6):1775–1777.
Roth, H. R., Lu, L., Seff, A., Cherry, K. M., Hoffman, J.,
Wang, S., Liu, J., Turkbey, E., and Summers, R. M.
(2014). A new 2.5 D representation for lymph node
detection using random sets of deep convolutional
neural network observations. In Medical Image Com-
puting and Computer-Assisted Intervention–MICCAI
2014, pages 520–527. Springer.
Shin, H.-C., Roth, H. R., Gao, M., Lu, L., Xu, Z., Nogues,
I., Yao, J., Mollura, D., and Summers, R. M. (2016).
Deep convolutional neural networks for computer-
aided detection: Cnn architectures, dataset charac-
teristics and transfer learning. IEEE transactions on
medical imaging, 35(5):1285–1298.
Urban, G., Bendszus, M., Hamprecht, F., and Kleesiek, J.
(2014). Multi-modal brain tumor segmentation using
deep convolutional neural networks. Proceedings of
BRATS-MICCAI.
Wang, J., MacKenzie, J. D., Ramachandran, R., and Chen,
D. Z. (2015). Neutrophils identification by deep learn-
ing and voronoi diagram of clusters. In Medical Im-
age Computing and Computer-Assisted Intervention–
MICCAI 2015, pages 226–233. Springer.
Wardlaw, J. M., Muir, K. W., Macleod, M.-J., Weir, C.,
McVerry, F., Carpenter, T., Shuler, K., Thomas, R.,
Acheampong, P., Dani, K., et al. (2013). Clinical rele-
vance and practical implications of trials of perfusion
and angiographic imaging in patients with acute is-
chaemic stroke: a multicentre cohort imaging study.
Journal of Neurology, Neurosurgery & Psychiatry,
pages jnnp–2012.
Wolterink, J. M., Leiner, T., Viergever, M. A., and I
ˇ
sgum,
I. (2015). Automatic coronary calcium scoring in car-
diac CT angiography using convolutional neural net-
works. In Medical Image Computing and Computer-
Assisted Intervention–MICCAI 2015, pages 589–596.
Springer.
Yang, D., Zhang, S., Yan, Z., Tan, C., Li, K., and Metaxas,
D. (2015). Automated anatomical landmark detection
on distal femur surface using convolutional neural net-
work. In Biomedical Imaging (ISBI), 2015 IEEE 12th
International Symposium on, pages 17–21. IEEE.
Zhao, L., Wu, W., and Corso, J. J. (2013). Semi-automatic
brain tumor segmentation by constrained mrfs using
structural trajectories. In International Conference on
Medical Image Computing and Computer-Assisted In-
tervention, pages 567–575. Springer.
Zikic, D., Glocker, B., Konukoglu, E., Criminisi, A., Demi-
ralp, C., Shotton, J., Thomas, O., Das, T., Jena, R.,
and Price, S. (2012). Decision forests for tissue-
specific segmentation of high-grade gliomas in multi-
channel mr. In International Conference on Medi-
cal Image Computing and Computer-Assisted Inter-
vention, pages 369–376. Springer.
Thrombus Detection in CT Brain Scans using a Convolutional Neural Network
33
