
Managing Provenance for Medical Datasets
An Example Case for Documenting the Workflow for Image Processing
Ajinkya Prabhune
1
, Rainer Stotzka
1
, Michael Gertz
2
, Lei Zheng
3
and J
¨
urgen Hesser
3,4
1
Institute for Data Processing and Electronics, Karlsruhe Institute of Technology, Eggenstein-Leopoldshafen, Germany
2
Database Systems Research Group, Institute of Computer Science, Heidelberg University, Heidelberg, Germany
3
Experimental Radiation Oncology, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany
4
Experimental Radiation Oncology, IWR, Heidelberg University, Heidelberg, Germany
Keywords:
Scientific Data Repository, ProvONE Provenance Standard, Open Annotation Data Model, DICOM Dataset,
Angioscopy Workflow, Metadata Management.
Abstract:
In this paper, we present a novel data repository architecture that is capable of handling the complex image
processing workflows and its associated provenance for clinical image data. This novel system has unique and
outstanding properties versus existing systems. Among the most relevant features are a flexible and intuitively
usable data and metadata management that includes the use of a graph-based provenance management strategy
based on a standard provenance model. Annotation is supported to allow for flexible text descriptors as being
widespread found for clinical data when structured templates are not yet available. The architecture presented
here is based on a modern database and management concepts and allows to overcome the limitations of
current systems namely limited provenance support, lacking flexibility, and extensibility to novel requests. To
demonstrate the practical applicability of our architecture, we consider a use case of automated image data
processing workflow for identifying vascular lesions in the lower extremities, and describe the provenance
graph generated for this workflow. Although presented for image data, the proposed concept applies to more
general context of arbitrary clinical data and could serve as an additional service to existing clinical IT systems.
1 INTRODUCTION
Electronic health records (EHR) offer a digital doc-
umentation of the diagnostic and therapeutic history
of a patient. Parts of these records are managed by
Hospital information systems (HIS) and subsystems
like radiology information systems (RIS). Over the
initiative integrating the healthcare enterprise (IHE)
defined workflows enable standardized records on one
hand side and (as a final goal) a complete coverage of
all procedures in a clinic. However, from the perspec-
tive of data provenance, these systems partially solve
the data management problem. Data provenance is
hereby of utmost relevance since it allows traceabil-
ity with respect to validation and reproducibility of
diagnostic and therapeutic procedures and decisions
(Estrella et al., 2007). However, despite its relevance,
this topic is scarcely discussed for clinical routine
data reporting with some exceptions in bioinformat-
ics (Davidson et al., 2007). Approaches that have
been widespread used are neither complete, nor flexi-
ble and are therefore difficult to integrate in a clinical
environment.
In the following, we propose a new architecture
for medical data repository system with a dedicated
focus on medical image data processing. Currently,
the image data is mostly obtained in radiology depart-
ments, and the acquired data such as Magnetic Reso-
nance Imaging (MRI) or Computerized Tomography
(CT) data is typically stored in a Picture Acquisition
and Communication System (PACS), backed by DI-
COM 3.0 format (Mildenberger et al., 2002). This
format allows storing image data and the associated
metadata such as patient id, type of acquisition, date,
etc, that is embedded within the DICOM files. Thus,
all relevant information required to repeat the acqui-
sition is stored and accessible via reading and inter-
preting the DICOM header files.
Although being in widespread use, two major lim-
itations of the PACS are, (a) the lack of a modern
database (NoSQL) technology for storing, indexing
and accessing metadata. (b) the brittleness of han-
236
Prabhune A., Stotzka R., Gertz M., Zheng L. and Hesser J.
Managing Provenance for Medical Datasets - An Example Case for Documenting the Workflow for Image Processing.
DOI: 10.5220/0006109402360243
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 236-243
ISBN: 978-989-758-213-4
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

dling the workflow and provenance information for
reproducing the results. For example, manipulations
of image data via image processing routines (which
will dominate in the future to automatically process
standard cases), the processed result is stored as new
DICOM data, even if only small modifications on the
header information were performed.
From the perspective of repeatability, the image
processing workflow would have to be stored in the
DICOM data as well. This can be accomplished either
by using private tags or by defining a new modality.
The former step is an ad-hoc solution since there are
no formal rules how this should be done and gener-
ally we cannot assume that all users are handling this
issue with sufficient care. Hence, there is no guaran-
tee that this image processing workflow can ever been
repeated. The latter strategy requires a new DICOM
modality standard, which is an immense overhead.
From this perspective, there is a demand for an
easy-to-use technology that not only maps all meta-
data formats, workflows, and patient related data but
also enables users to describe new workflows and still
guarantee the provenance information.
This lack of flexibility of the current system and
the brittleness of the DICOM standard led us to pro-
pose a different type of data repository architecture
that is overcoming these limitations. Instead of intro-
ducing a disruptive solution in the existing PACS in-
frastructure, we present an auxiliary system that pro-
vides the following functionality;
• Metadata management for extracting, modelling,
indexing and storing the metadata embedded from
DICOM-file in a flexible and a scalable database
• Provenance tracking using standard provenance
models such as ProvONE and PREMIS
1
for the
image processing workflows.
• Data annotation (image annotations) for systematic
capturing of vital details in the standard Open An-
notation Data Model (Sanderson et al., 2013).
• Data preservation for allowing long-term access
and reusability of the data.
• Data quality control and data curation for specific
tools controlling rules of good practice and later
diagnostic and treatment guidelines.
2 IMAGE PROCESSING DATA
REPOSITORY
The goal of the Image Processing Data Repository
(IPDR) is to provide the various auxiliary functional-
1
http://www.loc.gov/standards/premis/
!"#$%&#'#"()*+,'-*.
/%%*&0 &#*%(!$+1#"$2
30&0(3#2"*1$+4 567-#"0&#*%(!$+1#"$
8$&090 &0(:;&+0"&#*%
8$&090 &0(5+*"$22#%<
8$&090&0(8*9$--#%< 8$&090&0(!&*+0<$
30&0(5+$2$+10&#*%
30&0(/%0-42#2
30&0(=6+0&#*% 30&0(5+*"$22#%<
>53?(
!$+1#"$2
8$&09 0&0(
80%0<$@$%&
30&0(
80%0<$@$%&
/""$22(A04$ +(B /5>
30&0(C+0%2'$+
>%<$2&D3*.%-*09(
)*+,'-*.2
/6&E$% &#"0&#*%(0%9(
/6&E*+#F0&#*%
G02$(8$&090&0(
?$<#2&+0&#*%
>%&$+0"& #1$()$7(5*+&0-
H%*.-$9<$(?$I+$2$%&0&#*%
J2$+B
>%&$+'0"$
K5=(=-62&$ +
L/II-#"0&#*%(K*2&#%<(
!42&$@M
8$&090&0(!&*+0<$
30&0(/+"E#1$(
L5/=!(2$+1$+M
>@0<$(5+*"$22#%<(30&0(?$I*2#&*+4(
N$%$+#"(=-#$%&
Figure 1: Image Processing Data Repository Architecture.
ity for handling the lifecycle of DICOM dataset stored
in the PACS server. The IPDR follows a modular ar-
chitecture design, as shown in Figure 1. The modular
design of IPDR allows us to extend the functionality
to fulfil any new requirements put forth by the radia-
tion therapy (RT) research community. The complete
IPDR architecture is developed in Java, the various
REST services are implemented in Jersey (Sandoval,
2009) and the front-end is developed in Vaadin frame-
work. The IPDR is deployed on a web server (Tomcat
7). Following is the description of the core compo-
nents of the IPDR architecture.
2.1 Generic Client
The Generic Client component provides a convenient
solution for the RT medicine and research commu-
nity to integrate their existing software/tools with the
IPDR seamlessly. The Generic Client is an upgrade
to the GCS-API (Prabhune et al., 2015) that was pre-
viously developed for handling the datasets of the
nanoscopy research community. The Generic Client
is extended to handle the DICOM files. The various
modules of Generic Client are explained below:
• Access Layer-API: The access layer API exposes
the various interfaces for connecting to the existing
radiation therapy data processing tools. Currently,
we have developed a command line interface (CLI)
over the access layer API that allows the RT com-
munity to transfer the datasets.
• Base Metadata Registration: For long term
archival, it is necessary to register of the DICOM
files in IPDR. However, the DICOM files main-
Managing Provenance for Medical Datasets - An Example Case for Documenting the Workflow for Image Processing
237

tain its proprietary metadata schema that needs to
be translated to the standard metadata schema sup-
ported by the IPDR. This module enables the au-
tomated registration of the DICOM datasets by ex-
tracting the administrative metadata concerning the
DICOM Study, Series, and Image and mapping it
to the Core Scientific Metadata Model (CSMD)
(Matthews, 2015).
• Ingest/Download Workflows: Ingest/Download
workflows module holds the predefined workflow
that allow the ingestion of DICOM data from client
systems to the IPDR or to download the DICOM
datasets from the IPDR to the client system.
• Data Transfer: The data transfer module allows
the transfer of DICOM datasets bi-directional from
multiple endpoint, i.e. from client system or data
acquisition systems to IPDR or vise versa. Cur-
rently, the HTTP WebDAV (Goland et al., 1999)
protocol is supported for high-throughput transfer
of the datasets. The various interfaces for inte-
grating other transfer protocols such as, FTP and
gridFTP (Allcock et al., 2005) are available. The
data transfer module is designed to be fail-safe. For
example, data transfers that are interrupted are au-
tomatically re-triggered for transfer. To optimize
high-volume data transfers, the number of parallel
WebDAV connections can be manipulated.
• Authentication and Authorization: The Generic
Client follows a two-fold process. First, to enable
the registration of DICOM datasets , for a regis-
tered member of the RT research community, an
OAuth-secured RESTful connection is established.
Each researcher is provided with a unique access
token and access token secret. Second, for transfer-
ring the data, the WebDAV protocol authentication
is required. Currently, WebDAV authentication for
an entire research community is configured.
2.2 Image Processing Data Repository
The IPDR is a multi-module architecture that is sep-
arated into functionality specific module. The IPDR
is deployed as a web application with access to the
high-performance computing cluster, the large-scale
data storage, and a dedicated metadata database. Fol-
lowing is the description of each module of the IPDR:
IPDR Services: The services module provides the
various arbitrary high-end services for interacting
with datasets stored in IPDR.
• Scientific Workflow (medical Image Data Process-
ing workflow) submodule offers the integration
of a workflow engine for automating the execu-
tion of the image processing workflows. Cur-
rently, the Apache ODE
2
BPEL workflow engine
is integrated with IPDR and workflows defined
in BPEL specification (OASIS, 2007) are sup-
ported. The Prov2ONE (Prabhune et al., 2016) al-
gorithm handles the automatic creation of prospec-
tive ProvONE graph, which is stored in a graph
data model of the metadata storage.
• Data discovery submodule provides the users to
search the ingested DICOM datasets based on the
metadata stored in the metadata storage. The meta-
data are indexed for enabling full-text search, for
searching over provenance graphs various queries
implemented as RESTful services are provided.
Finally, to enable large scale metadata harvesting
the Open Archives Initiative Protocol for Metadata
Harvesting (OAI-PMH)
3
is implemented.
• Annotation service is an interactive submodule that
allows enriching of the images through the anno-
tation service. Valuable information provided in
form of annotations by the researchers is modeled
using the Open Annotation Data Model and main-
tained in the metadata storage.
• Publication service submodule allows users to
share (provide open access) the experiment dataset
with other research communities, which can be
based on data exchange technologies like i2b2
4
.
Metadata: The metadata module is responsible for
handling all the metadata specific tasks in the IPDR.
Metadata can either be embedded inside the DICOM
files or for other file formats it can be ingested sep-
arately with the datasets. For enabling querying,
sharing and reusing of metadata, DICOM metadata
needs to be stored in a dedicated metadata storage.
Furthermore, as DICOM metadata is subject to fre-
quent changes throughout its lifecyle, it is necessary
to have a flexible database model. Thus, we chose
ArangoDB
5
, a database which offers three different
types of data models, namely, key-value, document
and graph data model.
• Metadata Extraction: The various metadata ex-
tractors for extracting the metadata from DICOM,
HDF5, TIFF and XML files are provided as inde-
pendent micro-services by this module. For ex-
tending towards other file formats, this module
provides a generic interface that can be imple-
mented by any new metadata extractors.
• Metadata Modelling: The various metadata mod-
els (schemas) are registered through this com-
2
http://ode.apache.org/
3
https://www.openarchives.org/pmh/
4
www.i2b2.org
5
https://www.arangodb.com/documentation/
HEALTHINF 2017 - 10th International Conference on Health Informatics
238

ponent. For example, the ProvONE provenance
model, CSMD, Metadata Encoding and Transmis-
sion Standard (METS)
6
and PREMIS are some of
the currently supported metadata schemas. Fur-
thermore, application-specific metadata model of
DICOM is also registered by this submodule.
• Metadata Processing: The metadata processing
submodule provides the handling and assembling
of the community specified METS profile. METS
is metadata container format that comprises of var-
ious sections which allow encoding of adminis-
trative hamdSeci, structural hfileSeci, hstructMapi,
hstructLinki descriptive hdmdSeci and provenance
hdigiprovMDi metadata. The heterogeneous meta-
data comprising descriptive and administrative
metadata from the ArangoDB document store and
provenance metadata from graph store are assem-
bled in the METS profile for enabling sharing of
entire metadata for a dataset.
• Metadata Storage: The various database specific
storage adapter for storing and querying the meta-
data are implemented in this submodule. The
CRUD operations for document metadata repre-
senting the contextual information from the DI-
COM dataset, for graph metadata representing the
provenance and workflows, and the six verbs of
the OAI-PMH protocol are implemented in Arango
Query Language (AQL).
Data: The Data module provides the integration with
the low-level functionality that is responsible for stor-
ing the data in the cache storage and further in the tape
storage for long term archival. This data storage rep-
resents the PACS server, which is enriched with the
high-performance computing (HPC) cluster for en-
abling processing of the DICOM dataset, where these
data processing services (algorithms) are deployed on
the HPC. Following are functionalities that extend the
underlying PACS server.
• Data Preservation: The Data Preservation submod-
ule provides with the checksum of the dataset that
is to be ingested in IPDR. This checksum is main-
tained in the PREMIS metadata schema for verify-
ing the consistency of a file during data transfers
through Generic Client.
• Data Analysis and Curation: The various com-
munity specific data analysis and curation algo-
rithm are registered in these submodules. The data
processing algorithms are deployed as individual,
reusable processes that expose a unique REST end-
point, which is used when assembling a scien-
tific workflow (necessary for composing the BPEL
workflow).
6
http://www.loc.gov/standards/mets/
• Data Processing: The integration with the HPC
cluster and configuration of the execution envi-
ronment is handled by this submodule. More-
over, when new intermediary datasets are gener-
ated as a result of an execution of data processing
algorithm, the automated registration and ingest of
these datasets is performed by this submodule.
2.3 User Interface
A web-based user interface developed in Vaadin is
integrated with the IPDR. Feature such as free-text
search over metadata and faceted search are avail-
able for discovering the datasets. Provenance graphs
stored in ArangoDB are visually represented using the
D3.js (Zhu, 2013) framework. The OAI-PMH meta-
data services are exposed through the user interface
for harvesting the metadata. As the user interface
is integrated with the workflow engine, the radiation
therapy research community can perform remote exe-
cution of their image processing workflows.
3 DATA AND METADATA
WORKFLOW
Datasets in interventional radiological clinics follow
a systematic workflow, starting from the image acqui-
sition in DICOM format to the final generation of a
treatment record. During each step of the workflow,
the DICOM images are subject to image manipula-
tions and diagnostic information is extracted; these
datasets are hence enriched with essential metadata
describing the diagnostic details at each step. More-
over, additional and related datasets might be created
as well. To manage these datasets, we described the
complete flow of the DICOM datasets and metadata
beginning from the Data Acquisition system (DAQ)
and the treatment to making it accessible to clinicians
for finding similar cases or for quality assurance rea-
sons; and to use it for documentation. The complete
workflow, described in our example case consists of
eight steps as shown in Figure 2. (1) The workflows
begins when the either raw DICOM dataset acquired
from the CT scanner system is made available to the
Generic Client. (2) The Generic Client is entirely au-
tomated, it performs the registration of the dataset to
be ingested by extracting the base metadata from the
DICOM metadata section, translates it into the CSMD
standard and registers the dataset. (3) A successful
registration of base metadata triggers the transfer of
data to the PACS server. (4) The complete metadata
from the DICOM dataset is extracted, modelled and
Managing Provenance for Medical Datasets - An Example Case for Documenting the Workflow for Image Processing
239
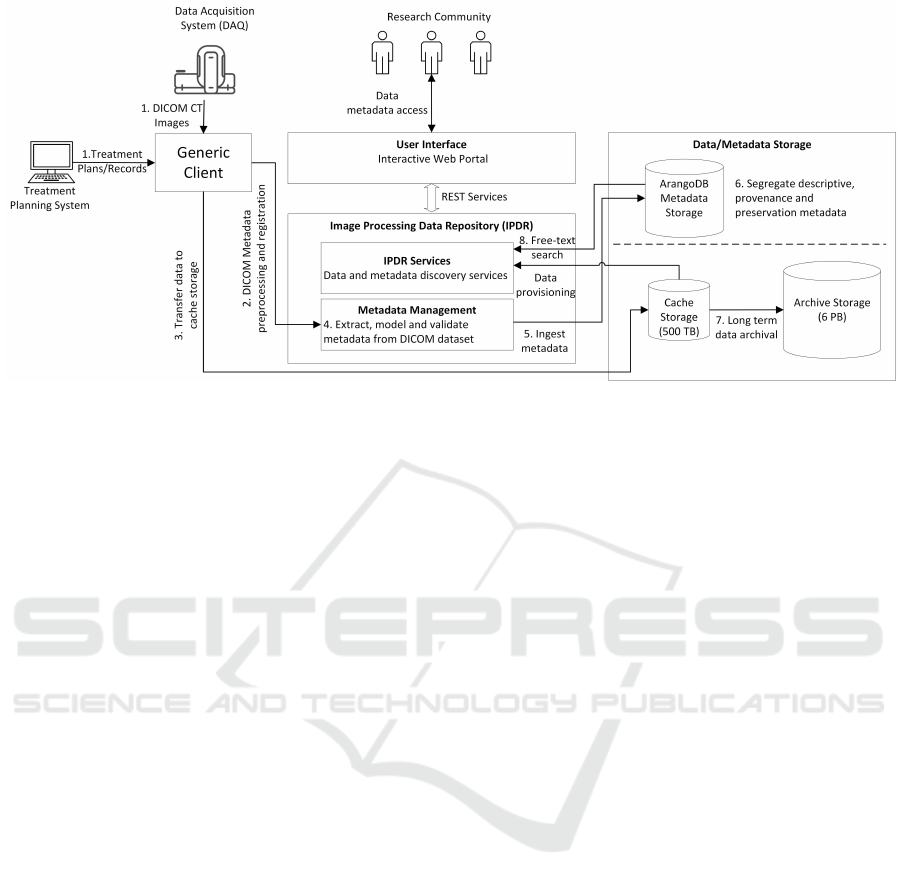
Figure 2: Flow of data and metadata from clinical data acquisition and treatment system to IPDR.
validated. (5) The metadata is ingested to the ded-
icated metadata storage database. (6) Metadata is
segregated as descriptive or provenance metadata and
stored either in document data model or graph data
model of ArangoDB. (7) The DICOM dataset from
the cache storage is transferred to archive storage for
long-term preservation. The preservation metadata
associated with the dataset is updated in the meta-
data storage. (8) The metadata is indexed for enabling
free-text search and allowing discovery of the datasets
from the IPDR. The data and metadata is accessible in
the Vaadin based user interface which is connected to
the IPDR through various REST services.
4 WORKFLOW AND
PROVENANCE
A successful completion of the Schedule (Acquisi-
tion) Workflow (SWF) generates the various DICOM
files that are stored in the PACS server. The Post-
Processing Workflow (PAWF) is the logical extension
of the SWF (Liu and Wang, 2010), which aims for de-
riving additional qualitative and quantitive data that is
beneficial for improving the patient’s treatment. Typ-
ically, in a clinical environment, there are two cate-
gories of post-processing workflows.
Distributed application (agent) oriented work-
flows: In the case of distributed agent oriented work-
flows, various information systems are controlled un-
der the authority of different health care actors such
as physicians, general practitioners and various hos-
pital departments. Currently, distributed agent ori-
ented post-processing workflow steps involves appli-
cations, such as Computer Aided Detection (CAD),
Image processing, 3D reconstruction and surface ren-
dering are available. However, these workflows are
often distributed, and the various applications partici-
pating in the execution of the workflow are deployed
on a stand-alone hosting system. Various endeavors
address the handling of provenance and workflows
in these distributed agent oriented workflows (Kifor
et al., 2006) (Zhang et al., 2009).
Custom data processing workflows: In the case of
custom data processing workflows the various data
processing steps (algorithms) are typically defined
and implemented for advanced data processing in the
clinical environment. Currently, to coordinate and
trace the execution of the workflow steps, the work-
list embedded in the DICOM file is referred. The
DICOM worklist (to-do lists) hold the list of tasks
that are to be performed on DICOM datasets. How-
ever, DICOM does not have the capability to model
and maintain the provenance traces associated with
the data. Even though the worklist offers a conve-
nient technique for maintaining the workflow steps,
it obviously lacks an accepted workflow and prove-
nance standard. For modelling comprehensive prove-
nance information, i.e. both prospective provenance
(workflow execution plan) and retrospective prove-
nance (runtime events) (Zhao et al., 2006), the W3C
ProvONE standard is adopted. Furthermore, to en-
able reuse of these data processing algorithms, they
are registered as web-services in the Data Analysis
and Curation sub-module of the IPDR. The interme-
diate data generated after each step in the workflow
is ingested in the IPDR PACS server, thus preventing
unnecessary repetition of the workflow.
Using an existing angioscopy image analysis
workflow (Maksimov et al., 2009) (Brockmann et al.,
2010), the various processing steps are deployed as
HEALTHINF 2017 - 10th International Conference on Health Informatics
240
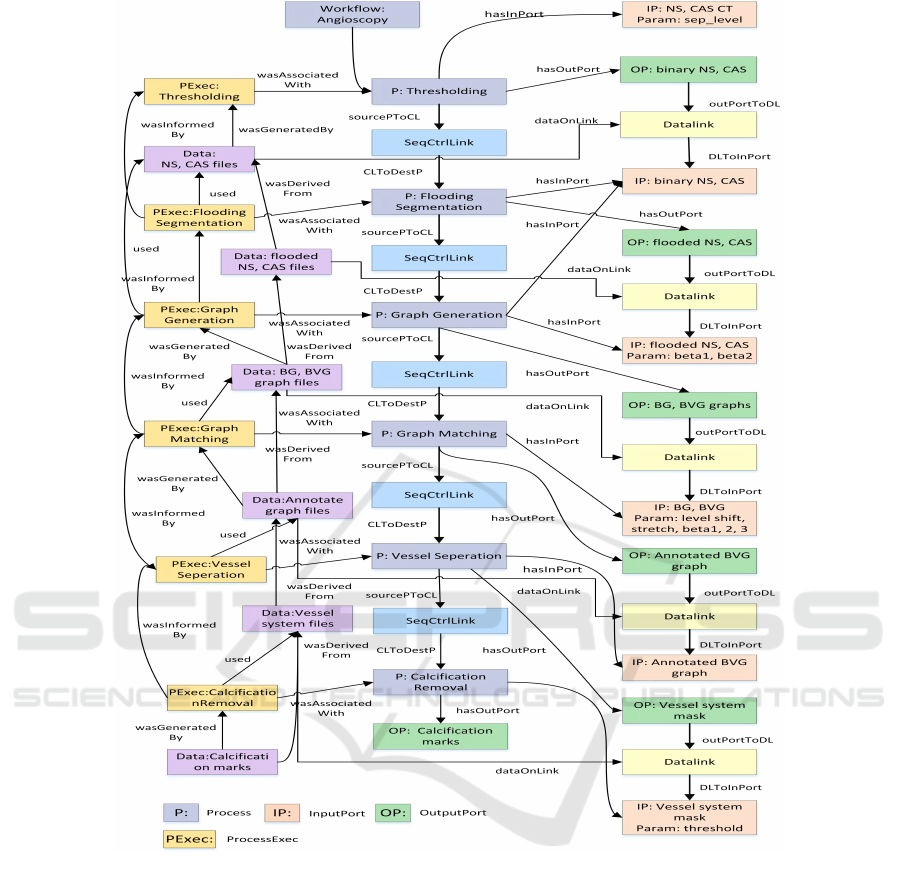
Figure 3: Angioscopy workflow provenance in ProvONE.
independent web services that are accessible through
a REST URI. Based on these REST URIs, the com-
plete workflow is modelled in BPEL specification and
executed on the high-performance computing cluster.
Before the execution of the workflow, the ProvONE
prospective graph is generated by the Prov2ONE al-
gorithm, and during the execution enriched with the
runtime provenance information (retrospective prove-
nance). The complete ProvONE provenance graph for
the angioscopy workflow is shown in Figure 3.
During the execution of the image analysis work-
flow the intermediate result are ingested into the IPDR
and the metadata is extracted, modelled and stored in
the metadata storage, as shown in the Figure 2.
5 DISCUSSION
In the available systems, clinical process data is stored
in diverse clinical information systems, for example
in HIS including subsystems such as a RIS and PACS.
The essence of these systems is not only the reporting
but also to be able to retrospectively follow the course
of the clinical decision and treatment. Whenever im-
age data is manipulated such as via image processing
or conclusions are driven via machine learning tech-
niques, there is no adequate tool available yet that al-
lows tracking these steps in an adequate way. The
rationale for considering the IPDR is its high flexibil-
ity for different clinical processes and their adequate
Managing Provenance for Medical Datasets - An Example Case for Documenting the Workflow for Image Processing
241

documentation.
Furthermore, the benefit of being able to track pre-
vious diagnostic and treatment processes is to be able
to assess the treatment outcome or the correctness of
the diagnostic step. This allows both to increase the
internal quality of clinical routines but also offers the
chance to get a deeper insight when e.g. for cancer
patients there is a relapse, which might offer different
ways of further patient treatment.
However, to handle the frequent metadata changes
in the DICOM dataset that are introduced during the
clinical studies, we integrated a database system sup-
porting multiple flexible data models. Additionally,
the metadata is automatically indexed for enabling
free-text search through the data discovery service.
For capturing the dynamic information in the form
of annotations, the W3C standard Open Annotation
Data Model is implemented through the annotation
service. The IPDR allows the entire modelling of
the DICOM metadata in METS format and the ret-
rospective provenance is modelled in PREMIS stan-
dard, thus allowing efficient sharing (harvesting) of
the metadata using the OAI-PMH services. For han-
dling the provenance of the image processing work-
flows, we integrated the ProvONE standard in IPDR,
wherein both the workflow plan as well as the asso-
ciated provenance are captured, thus, enabling repro-
ducibility of scientific results.
We also presented the data and metadata workflow
describing the integration of IPDR with the PACS
server, see Figure 2. Our aim was to seamlessly in-
tegrate the entire IPDR into the existing PACS server
and network infrastructure without any disruption in
the existing execution environment. The DICOM
dataset either from the data acquisition system (imag-
ing modalities) or from the various workstations in the
PACS network is processed through IPDR and for-
warded to the PACS server. To handle the DICOM
data the open-source DICOM DCM4CHE (Zeilinger
et al., 2010) toolkit is integrated into the IPDR.
Regarding the regulations, documents concerning
diagnostic and therapeutic procedures have to be filed
for about 10-30 years, documents for quality control
have to be archived for about 5 years depending on
country and regulations. There is no formal require-
ment for a detailed documentation of medical pro-
cesses so far but the availability of a flexible prove-
nance software technique would foster regulation pro-
cedures if publicly available.
6 RELATED WORK
Currently, there are various commercial as well as
custom implementations of PACS solutions available
for clinical studies for handling the medical datasets.
Enterprise Imaging Repository (EIR) (Bian et al.,
2009) is an alternative to the commercial PACS server
and network solution that provides handling of DI-
COM files using the DCM4CHE toolkit. Various
functionality like HL7(Dolin et al., 2006) interfaces,
web access to DICOM objects and media creation ser-
vices (CDW) are implemented by the EIR.
For handling the cardiology datasets (Marcheschi
et al., 2009) have a PACS solution completely built
using open-source technology. This solution is based
on existing commodity hardware with a server farm
comprising a storage pool of 24 TB for saving the
cardiology datasets. A PACS based solution with au-
tomated workflow management techniques using the
YAWL specification in radiology information system
(Zhang et al., 2009) are implemented for allowing
a workflow-aware and flexible integration of various
components in building RIS.
The integration of a workflow system does of-
fer novel approach for a flexible RIS design. How-
ever, the above mentioned PACS solutions fail to
provide many of the critical aspects associated with
the handling of the complete lifecycle of the med-
ical datasets. The IPDR solution presented in this
paper overcomes these limitations by providing func-
tionalities such as, metadata handling based on stan-
dard metadata models, dedicated scalable and flexible
metadata storage, annotation services based on stan-
dard Open Annotation Data Model, integration with
high-performance computing cluster for performant
execution of post-processing workflow tasks and cap-
turing of provenance in ProvONE model.
7 CONCLUSION
In this paper, we presented the detailed architecture
and the functionalities provided by the IPDR, which
is a comprehensive repository system for handling the
EHR datasets. The IPDR is based on the principle
of modular architecture that enables easy extensibility
by adding task-specific modules for handling any new
requirements. The functionality provided by IPDR
enables the RT researchers to: (a) automatically ex-
tract, store, and access metadata in standard metadata
models, (b) allow reproducibility of scientific results
by capturing provenance in ProvONE standard, (c)
enrich the data quality by capturing annotations in the
Open Annotation Data Model, (d) enable the repeata-
HEALTHINF 2017 - 10th International Conference on Health Informatics
242

bility of complex image processing workflows using
integrated workflow engine, (e) large-scale metadata
harvesting through standard OAI-PMH protocol.
To demonstrate the handling of image processing
workflow with capturing of its associated provenance,
the angioscopy workflow modelled in BPEL was ex-
ecuted using a workflow engine integrated with the
IPDR, and the associated provenance was automati-
cally modelled in ProvONE (Figure 3).
However, the data exchange approach uses safe
data transfer strategies but are not yet adapted to clin-
ical needs. An integration of i2b2 and a data ware-
house that collects all clinical data in combination
with technologies such as PCORnet
7
offers the poten-
tial to standardised data exchange for studies and con-
sultation and will thereby allow using our architecture
to be integrated into a clinic-wide information system
of the next generation. In particular, this strategy is
considered to be integrated into data integration cen-
tres that are currently planned at the university med-
ical centre Mannheim (UMM) and the MIRACUM
8
consortium that is currently funded by the German
Ministry of Research and Education (BMBF). The
technology is built upon developments of the BMBF-
project LSDMA
9
.
REFERENCES
Allcock, W., Bresnahan, J., Kettimuthu, R., et al. (2005).
The globus striped gridftp framework and server. In
Proceedings of the 2005 ACM/IEEE conference on
Supercomputing, page 54. IEEE Computer Society.
Bian, J., Topaloglu, U., and Lane, C. (2009). Eir: En-
terprise imaging repository, an alternative imaging
archiving and communication system. In 2009 Annual
International Conference of the IEEE Engineering in
Medicine and Biology Society, pages 2168–2171.
Brockmann, C., Jochum, S., Hesser, J., et al. (2010). Graph-
matching-based computed tomography angiography
in peripheral arterial occlusive disease. Clinical Imag-
ing, 34(5):367 – 374.
Davidson, S. B., Boulakia, S. C., Eyal, A., Lud
¨
ascher, B.,
McPhillips, T. M., Bowers, S., Anand, M. K., and
Freire, J. (2007). Provenance in scientific workflow
systems. IEEE Data Eng. Bull., 30(4):44–50.
Dolin, R. H., Alschuler, L., Boyer, S., Beebe, C., Behlen,
F. M., Biron, P. V., and Shabo (Shvo), A. (2006).
Hl7 clinical document architecture, release 2. Jour-
nal of the American Medical Informatics Association,
13(1):30–39.
Estrella, F., Hauer, T., McClatchey, R., Odeh, M., Rogulin,
D., and Solomonides, T. (2007). Experiences of en-
7
www.pcornet.org
8
www.miracum.de
9
www.helmholtz-lsdma.de
gineering grid-based medical software. International
Journal of Medical Informatics, 76(8):621 – 632.
Goland, Y., Whitehead, E., Faizi, A., Carter, S., and Jensen,
D. (1999). Http extensions for distributed authoring–
webdav. Technical report.
Kifor, T., Varga, L. Z., Vazquez-Salceda, J., Alvarez, S.,
Willmott, S., Miles, S., and Moreau, L. (2006). Prove-
nance in agent-mediated healthcare systems. IEEE In-
telligent Systems, 21(6):38–46.
Liu, Y. and Wang, J. (2010). PACS and digital medicine:
essential principles and modern practice. CRC Press.
Maksimov, D., Hesser, J., Brockmann, C., Jochum, S., Di-
etz, T., et al. (2009). Graph-matching based cta. IEEE
Transactions on Medical Imaging, 28(12):1940–1954.
Marcheschi, P., Ciregia, A., Mazzarisi, A., Augiero, G., and
Gori, A. (2009). A new approach to affordable and re-
liable cardiology pacs architecture using open-source
technology. In 2009 36th Annual Computers in Car-
diology Conference (CinC), pages 537–540.
Matthews, B. (2015). Csmd: the core scien-
tific metadata model. Online http://icatproject-
contrib.github.io/CSMD/csmd-4.0.html.
Mildenberger, P., Eichelberg, M., and Martin, E. (2002).
Introduction to the dicom standard. European Radiol-
ogy, 12(4):920–927.
OASIS (2007). Standard, O.A.S.I.S: Web services busi-
ness process execution language version 2.0. Online
http://docs.oasis-open.org/wsbpel/2.0/OS/wsbpel-v2.
Prabhune, A., Stotzka, R., Jejkal, T., Hartmann, V., Bach,
M., Schmitt, E., Hausmann, M., and Hesser, J. (2015).
An optimized generic client service api for managing
large datasets within a data repository. In Big Data
Computing Service and Applications (BigDataSer-
vice), 2015 IEEE First International Conference on,
pages 44–51.
Prabhune, A., Zweig, A., Stotzka, R., Gertz, M., and
Hesser, J. (2016). Prov2ONE: An Algorithm for
Automatically Constructing ProvONE Provenance
Graphs, pages 204–208. Springer Publishing.
Sanderson, R., Ciccarese, P., Van de Sompel, H., Bradshaw,
S., Brickley, D., a Castro, L. J. G., et al. (2013). Open
annotation data model. W3C community draft.
Sandoval, J. (2009). Restful java web services: Master core
rest concepts and create restful web services in java.
Packt Publishing Ltd.
Zeilinger, G., Montgomery, O., Evans, D., et al. (2010).
The dcm4che project. Online Sourceforge project
https://sourceforge.net/projects/dcm4che/.
Zhang, J., Lu, X., Nie, H., Huang, Z., and van der Aalst,
W. M. P. (2009). Radiology information system: a
workflow-based approach. International Journal of
Computer Assisted Radiology and Surgery, 4(5):509–
516.
Zhao, Y., Wilde, M., and Foster, I. (2006). Applying
the Virtual Data Provenance Model, pages 148–161.
Springer Berlin Heidelberg, Berlin, Heidelberg.
Zhu, N. Q. (2013). Data visualization with D3. js cookbook.
Packt Publishing Ltd.
Managing Provenance for Medical Datasets - An Example Case for Documenting the Workflow for Image Processing
243
