
Collaborative Reproducible Reporting
Git Submodules as a Data Security Solution
Peter E. DeWitt
1
and Tellen D. Bennett
2
1
Biostatistics and Bioinformatics, Colorado School of Public Health,
University of Colorado Denver, Anschutz Medical Campus, Aurora, CO, U.S.A.
2
Pediatric Critical Care, University of Colorado School of Medicine, Children’s Hospital Colorado, Aurora, CO, U.S.A.
Keywords:
Data Security, Collaborative Authoring, Reproducible Reports, Workflow, Software.
Abstract:
Sensitive data and collaborative projects pose challenges for reproducible computational research. We present
a workflow based on literate programming and distributed version control to produce well-documented and
dynamic documents collaboratively authored by a team composed of members with varying data access priv-
ileges. Data are stored on secure institutional network drives and incorporated into projects using a feature
of the Git version control system: submodules. Code to analyze data and write text is managed on public
collaborative development environments. This workflow supports collaborative authorship while simultane-
ously protecting sensitive data. The workflow is designed to be inexpensive and is implemented primarily
with a variety of free and open-source software. Work products can be abstracts, manuscripts, posters, slide
decks, grant applications, or other documents. This approach is adaptable to teams of varying size in other
collaborative situations.
1 INTRODUCTION
Reproducible reporting, defined here as processing
data and generating an abstract, manuscript, slide
deck, or poster via a fully documented and auto-
mated process, is considerably more difficult when
working with multiple authors and sensitive data,
such as protected health information (PHI). Work-
flows for reproducible computational research using
tools such as the Jupyter Notebook
1
, the Galaxy
project
2
, or RStudio (Gandrud, 2015) are not con-
sistently used in biomedical research (Peng et al.,
2006; National Academies of Sciences, Engineering,
and Medicine, Division on Engineering and Physical
Sciences, Board on Mathematical Sciences and Their
Applications, Committee on Applied and Theoretical
Statistics, 2016). This may be due to concerns about
slower production, the need for investigators to learn
new tools, or barriers to collaboration between inves-
tigators with varying computational skills and devel-
opment environments. Collaborative research involv-
ing sensitive data poses additional challenges.
One solution would be for a team to work in a
single development environment hosted on a compu-
1
http://jupyter.org/
2
http://galaxyproject.org/
tational server with the necessary physical and elec-
tronic security standards for the level of sensitivity of
the data. However, the financial investment required
to build a full development environment behind an in-
stitutional firewall might be prohibitive for some re-
search teams. Fortunately, a reproducible collabora-
tive workflow that protects sensitive data is possible
at much lower cost.
We minimize team hardware and software ex-
penses in two ways. First, only those team mem-
bers who require data access are provided with
institutionally-owned laptops with licenses for whole-
disk encryption and other proprietary software. Sec-
ond, by using free and open-source software for ver-
sion control, analysis, and manuscript authoring, we
incur minimal financial expenses when new team
members join or when we collaborate with external
investigators.
Our solution to data protection and collabora-
tion is to compartmentalize and distribute our project
such that data resources, analysis scripts, and text
are all linked together, version-controlled, and access-
controlled via implicit and explicit read/write permis-
sions. Raw data is stored on institutionally owned
network drives and cloned on laptop computers which
have been approved for storage of our data. Only team
230
Dewitt P. and Bennett T.
Collaborative Reproducible Reporting - Git Submodules as a Data Security Solution.
DOI: 10.5220/0006109302300235
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 230-235
ISBN: 978-989-758-213-4
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

members with institutional review board approval can
access the data. Data analysis scripts and manuscript
text files are available to all team members on pub-
lic code hosting services. The linkage between the
data and the code is made possible by a feature of
the Git version control software: submodules. A 40-
character hexadecimal sequence (SHA-1 hash) allows
us to share the version of the data source publicly
without compromising the data itself.
The objective of this manuscript is to present a
workflow that we developed to 1) protect sensitive
data from unauthorized access, 2) allow multiple au-
thors, included those with and without data access
rights, to contribute to a single set of files, and 3) min-
imize the financial commitment to hardware and soft-
ware.
Our primary focus is on the use of the Git version
control system and specifically, Git submodules. We
will note other software tools and programs used in
our workflow, but they can often be substituted for
other similar software.
2 WORKFLOW OVERVIEW
Dynamic document authoring is a key component of
the overall reproducible research paradigm. Varia-
tions on literate programming (Knuth, 1984) are ideal
for this purpose. The R package knitr (Xie, 2015),
an evolution of R’s sweave (Leisch, 2002) pack-
age, provides a structured process for authoring a
manuscript using a literate programming paradigm.
knitr was highlighted several times at a recent
workshop supported by the National Academies
of Sciences, Engineering, and Medicine (National
Academies of Sciences, Engineering, and Medicine,
Division on Engineering and Physical Sciences,
Board on Mathematical Sciences and Their Applica-
tions, Committee on Applied and Theoretical Statis-
tics, 2016).
We typically perform data analysis with the sta-
tistical language R
3
and rely on either markdown
or L
A
T
E
X for markup. The desired format of our
deliverables dictates the markup language selection.
Weaving R code with a markup language is well-
described (Gandrud, 2015).
Our team manages collaborative projects using a
distributed version control system, Git
4
. Git is free
to use and is supported on all major operating sys-
tems. Distributed version control systems are becom-
ing more common than centralized systems, although
3
https://www.r-project.org/
4
https://Git-scm.com/
some distributed version control projects, including
many of ours, have a centralized design (De Alwis
and Sillito, 2009).
In the simplest centralized design, a Git server
hosts the repository and each team member would
push to, and pull from, that server. It is possible to
have the individual team members’ repositories di-
rectly linked, but we did not use this option because
of network security concerns. Another option is to
have a bare repository on a network drive act as the
central code repository. We use that design for a mi-
nority of projects with unusually sensitive data. For
most projects, our team takes advantage of the inte-
grated issue tracker, web editing interface, and addi-
tional read/write permissions provided by a Git server.
Several public Git repository sites exist. We chose
to use Atlassian’s Bitbucket
5
to host our reposito-
ries. At the time this choice was made, Bitbucket
allowed academic account holders unlimited private
repositories and unlimited collaborators. Recently,
Github.com has offered similar packages.
Code repositories solved the problems of dynamic
document authoring and collaboration, but we also
needed to track data set versions and limit data ac-
cess to approved team members without preventing
collaboration.
The solution was to use Git submodules. “Sub-
modules allow you to keep a Git repository as a subdi-
rectory of another Git repository. This lets you clone
another repository into your project and keep your
commits separate.” (Chacon and Straub, 2014). Also,
while the data files within the submodule exist in a
subdirectory and are visible in the working directory,
only the SHA-1 of the commit of the submodule is
stored in the primary project repository. Thus, when
the manuscript repository is pushed to bitbucket.org,
the only reference to the data is a 40-digit hexadec-
imal number. The data never leaves the team mem-
bers’ machines, but the status of the data is shared
and documented between team members.
3 INFRASTRUCTURE
Below we describe how we have used existing in-
frastructure, open source software, and free hosting
services to generate reproducible reports while pro-
tecting sensitive data. We designed the workflow
so that sensitive data is stored on a secure network
hard drive or whole-disk encrypted personal machine.
Data transfer between the network drive and a team
member’s machine only occurs on the institution’s
5
https://bitbucket.org
Collaborative Reproducible Reporting - Git Submodules as a Data Security Solution
231

network. The following subsections describe the nec-
essary hardware, repository design, and workflow for
collaboration.
3.1 Hardware
Our institution maintains a Microsoft Windows net-
work. We chose to work on Windows machines be-
cause they are available to all of our team members
and because they support whole-disk encryption soft-
ware that meets our institution’s requirements for data
security. Each team member with access to the data
has a whole-disk encrypted laptop or desktop. This
software costs approximately 100 US Dollars per ma-
chine, but allows each team member to have local
copies of the data repositories relevant to their work.
Like investigators at many academic institutions, we
have access to secure network drives behind the uni-
versity’s firewall. We rely on the network drives for
data repository hosting and backup.
3.2 Repository Design
Although Git is a distributed version control platform,
we conceptually have central data and code reposito-
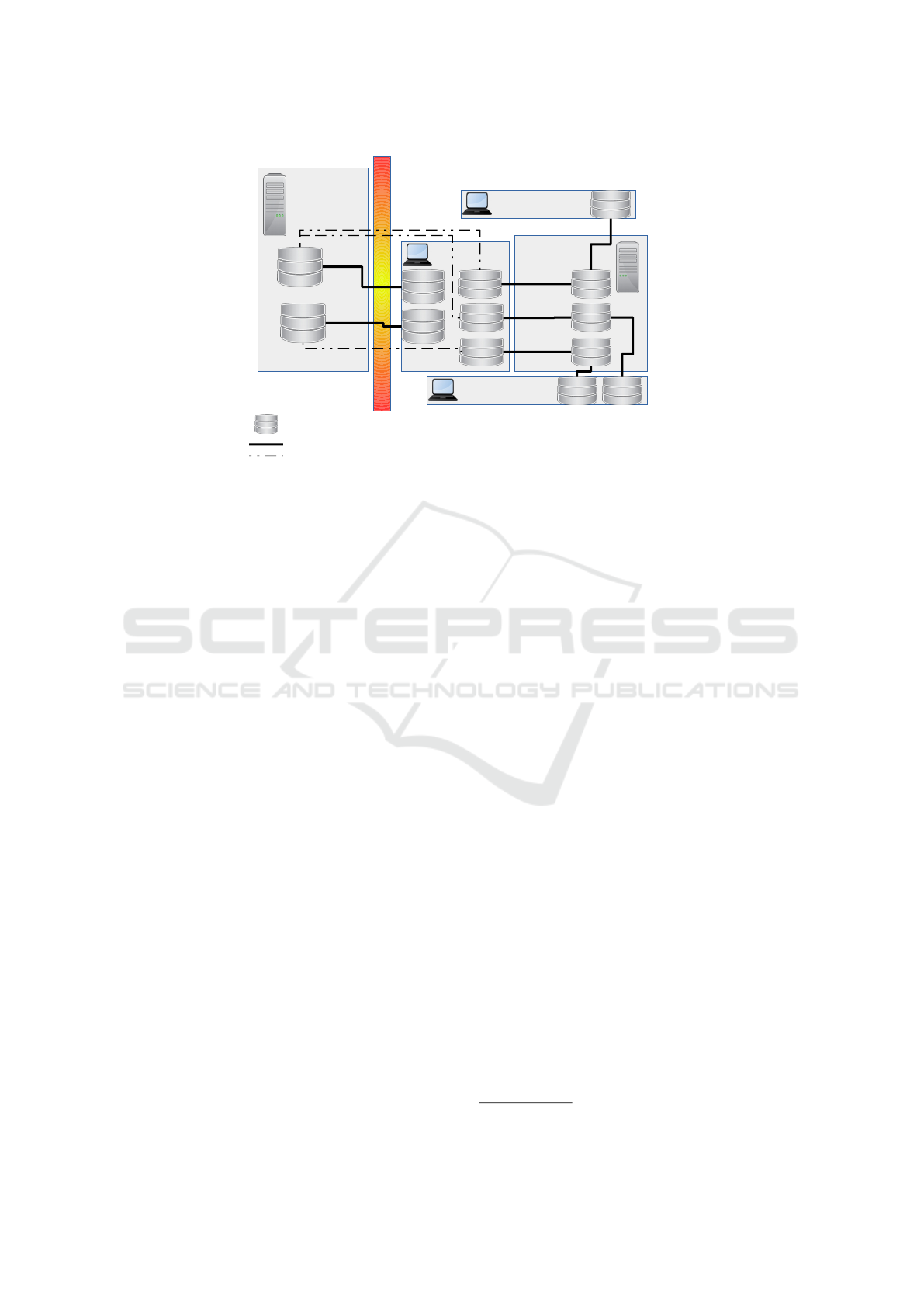
ries on a network drive or Git server, see Figure 1.
Each collaborator has a local clone of the necessary
data and code repositories on their machine that serve
as distributed backups of the central data and code.
3.2.1 Data Repositories
Data are housed in .csv format within our local data
repositories. For collaboration, team members with
data access privileges push to and pull form bare Git
repositories on our institution’s secure network drives.
Bare repositories do not contain a working directory:
individual files are not visible when inspecting the
contents of the directory and subdirectories. As such,
inadvertently editing or over-writing the data files is
very unlikely. We rely on the read/write access limits
enforced by the institution’s network to limit access to
these bare repositories and entrust those with access
to not manually edit files. The repositories theoret-
ically could become corrupted. If that occurred, we
would compare the distributed copies of the repos-
itories between team members and re-initialize the
repository from the most current local copy. This is an
advantage of the distributed version control paradigm:
every local copy is a backup for all others.
3.2.2 Code Repositories
In the simplest form of this workflow, a work product
such as a manuscript has its own code repository.
A basic repository design shown in Figure 2. An
example code repository can be found at https://
bitbucket.org/pedstbi/example collaborative report
which has a data submodule available at https://
bitbucket.org/pedstbi/example collaborative data
source. The analysis and manuscript authoring
code is free of sensitive data. Therefore, the remote
code repository can be maintained on a publicly
available code development system such as GitHub
or Bitbucket. We use private code repositories to
maintain academic confidentiality prior to manuscript
publication. Repositories on either GitHub or Bit-
bucket can be made public at any time, such as when
a manuscript is submitted for publication.
A team member working on a project (manuscript,
in this case) would have a local clone of the code
repository on their machine. Their daily workflow
would be to fetch and merge (pull-ing is shorthand for
the fetch then merge process) any changes on the re-
mote repository made by other team members, make
changes to files using the text editor of their choice,
stage and commit the changes using their local Git in-
stance, then push those changes to the remote reposi-
tory. Team members (clinical authors or collaborators
at other institutions, for example) whose contributions
are focused on writing the manuscript or who do not
have a whole-disk encrypted machine might have a
local copy of the code repository but not the data
repository. Those team members can have the bene-
fits of a version-controlled project without cloning the
data submodule. One challenge introduced by this ap-
proach is that collaborators without local data reposi-
tories cannot compile manuscripts. Because the quan-
titative results in the manuscript are generated by em-
bedded analytic code within the manuscript file, those
results cannot be updated without a local data copy.
Periodically, team members with both data and code
access must compile the manuscript (which runs the
embedded code) and commit the finished product to
the central code repository for reference by collabo-
rators who primarily write and edit manuscript text.
3.2.3 Limitations
The size of data submodules is the most important
limitation of this repository design. Thus far, the
largest data submodule in our system is approxi-
mately 10GB. Segmentation of the data repositories
into, for example, a large raw data repository and a
smaller analysis data repository for one project can
improve efficiency.
Additional features of Git such as branch-ing
strategies, forking, pull requests, rebase-ing, and oth-
ers, provide additional levels of structure within the
collaboration. However, such tools can be over-
HEALTHINF 2017 - 10th International Conference on Health Informatics
232
F
i
r
e
w
a
l
l
Shared
Network
Drive
Developer
Hosting
Service
DS1
DS2
DS1
DS2
M3
M2
M1
Collaborator 1
M1
Collaborator 2
M3 M2
M1
M3
M2
Repository (DS: Data Set; M: Manuscript)
F
i
r
e
w
a
l
l
Direct remote access
Access only through submodules
Figure 1: Collaboration Structure. Data is version-controlled in bare repositories on our institutional shared network drives.
The datasets are tracked within projects as submodules. Each developer has access to the data on his or her whole-disk
encrypted laptop or desktop. Non-sensitive code, manuscript text, references, etc. are hosted on bitbucket.org. Other authors
are able to contribute by having access to the bitbucket.org code repositories. Note that the manuscript repositories, M1,
M2, and M3, only have access to the data sets via git submodules. The copies of M1, M2, and M3 on the hosting service
and on each collaborator machine have no access to the data sets. The hosting service and collaborator only see a 40-digit
hexadecimal SHA1 to reference the version of the data repository.
whelming for a novice Git user. Increased train-
ing time, or limited participation, must be weighed
against the benefit of Git feature use.
This workflow does include copies of sensitive
datasets on the whole-disk encrypted local machines
of selected team members. Our experience has been
that data owners and institutional review boards are
supportive of this approach. If a particular dataset was
not permitted to be housed on a local machine with
whole-disk encryption, then a computational server
within the institution’s firewall would likely be nec-
essary.
Clinical members of our research team without
computational backgrounds have been able to adopt
most or all of this workflow with a modest time in-
vestment. However, like all complex tools, regular
use is needed to maintain comfort. A more integrated
environment that was friendly to the na
¨
ıve user would
increase the accessibility of a reproducible reporting
workflow.
3.2.4 Extensions/Other Options
Our team initially hosted code repositories on GitHub
and moved to Bitbucket as the team grew and the
number of projects increased. GitLab.com is another
option that offers unlimited private repositories, un-
limited collaborators, and up to 10GB disk space per
repository (compared to Bitbucket’s 1GB soft and
2GB hard limits). Placing a dedicated Git server be-
hind our institutional fire wall would provide a solu-
tion for data management and access control and use-
ful collaboration tools. Hardware and administrative
support costs would need to be considered.
4 COSTS
This reproducible reporting workflow is powerful and
also cost-effective. For many investigators, a Win-
dows operating system and Windows Office software
are supported by the institution. Whole-disk encryp-
tion software is inexpensive (100 US Dollars per team
member). Other software needed to implement this
workflow is free to use under the GNU General Pub-
lic License (GPL)
6
or similar license. There are no
hardware costs if investigators currently have indi-
vidual computers capable of performing the planned
analyses and access to a secure network drive. Many
academic investigators already have this hardware in
place.
The time and effort needed to learn the necessary
tools to adopt this workflow are likely higher than
the software and hardware costs. However, the re-
turn on investment can be high. Our experience in an
academic research environment suggests that a team
6
http://www.gnu.org/licenses/gpl-3.0.en.html
Collaborative Reproducible Reporting - Git Submodules as a Data Security Solution
233

. <user-path>/project1/
|-- .git/ # the Git repository
|-- analysis-scripts/ # data analysis scripts
| |-- data-import.R
| |-- primary-analysis.R
| |-- secondary-analysis.R
| ‘- figures.R
|-- data/ # A Git submodule
|-- products_donotedit/ # generated files
| |-- cache/
| | |-- documentation-data-import-cache/
| | |-- documentation-analysis-cache/
| | ‘-- manuscript-cache/
| |-- figures/
| |-- tables/
| |-- coverletter.docx
| |-- coverletter.md
| |-- documentation-data-import.html
| |-- documentation-analysis.html
| |-- manuscript.docx
| |-- manuscript.md
| ‘-- poster.pdf
|-- coverletter.Rmd ## Files for authoring
|-- documentation-data-import.Rmd ## coverletters,
|-- documentation-analysis.Rmd ## documentation,
|-- manuscript.Rmd ## manuscripts, posters,
|-- poster.Rnw ## etc.
‘-- README.md # project README
Figure 2: A generic repository layout for a manuscript writ-
ten in Rmarkdown. Not shown in the graphic, but part of
our overall design, are build scripts. A build script is a R
script, .cmd or .sh file, or makefile. The format and loca-
tion of the build script is project-specific. We decide which
format to use based on the complexity of the build required,
the development platforms (Windows, Mac, or Linux), the
integrated development environments (RStudio or vim are
used by our team), and ease of use.
adopting this workflow might see research production
slow for up to six months, recover to initial levels
within a year, and show potential increases after one
year. Improvements in quality and reproducibility are
difficult to quantify but are valuable.
5 ALTERNATIVE APPROACHES
Another solution to the simultaneous problems of
multiple collaborators and sensitive data might be
to run a local instance of Galaxy.
7
However, most
Galaxy tools use Python.
8
Few clinical researchers
have the training and experience to collaboratively
develop analysis code in Python. Many more have
been trained to use R. A capable computational server
would solve the problems of multiple collaborators
and data security. However, the purchase (5,000 US
7
https://galaxyproject.org/
8
https://www.python.org/
Dollars and up) and maintenance (varying, but poten-
tially exceeding 1, 000 US Dollars per year) costs for
such a server are beyond the reach of most small re-
search teams. Because many biomedical manuscripts
are generated by small teams, we think it likely that
the workflow we present here will be generalizable.
Existing cloud-based solutions such as RunMy-
Code.org
9
and the Open Science Framework
10
are re-
producible and support multiple collaborators, but are
not designed to protect sensitive data. Cloud-based
computational server services, some of which now
have robust data security features, are another op-
tion. Their utility will grow once institutional review
boards and data owners (health care organizations, in-
surance companies, etc.) gain enough confidence in
the data security measures used by those services that
researchers are consistently permitted to analyze sen-
sitive datasets in those environments.
We did not extensively test our Git-based solution
against other possible solutions. This was primarily
for two reasons. First, most available alternative ap-
proaches did not provide sufficient data security. Sec-
ond, alternative approaches with sufficient data secu-
rity required additional financial commitment beyond
standard operating expenditures. We developed this
workflow as part of an active academic research team
and needed to maintain productivity in our content
areas. The lack of formal method comparison is a
limitation of this manuscript at this time. However,
our team’s ability to rapidly adopt this workflow and
maintain productivity highlights the value and ease of
use of this approach.
6 DISCUSSION
Collaborative and reproducible biomedical reporting
can be inexpensive and have low barriers to entry
even when working with sensitive data and a team
with variable technical skills. Our goal is to intro-
duce an overall workflow and one set of viable tools.
Many data processing/analysis languages, markup
languages, text editors, file formats, and file sharing
systems can be used.
Peng (Peng et al., 2006; Peng, 2011) has sug-
gested criteria for the reproducibility of epidemio-
logic and clinical computational research. The work-
flow we present here would meet the criteria for Meth-
ods (free, open-source software, public code reposi-
tories), Documentation (well-commented code in the
repository), and Distribution (code repositories on
9
http://www.runmycode.org/
10
https://osf.io/
HEALTHINF 2017 - 10th International Conference on Health Informatics
234

public Git servers). However, due to the limitations
regarding disclosure of data, our workflow would not
meet Peng’s Data Availability criterion. Summary
statistics (Peng et al., 2006) could in some situations
be posted publicly, but overall the balance between
reproducibility and data privacy will need additional
public discussion (National Academies of Sciences,
Engineering, and Medicine, Division on Engineering
and Physical Sciences, Board on Mathematical Sci-
ences and Their Applications, Committee on Applied
and Theoretical Statistics, 2016).
Rossini and Leisch described how “information
and knowledge was divided asymmetrically between
[collaborators]. . . ” (Rossini and Leisch, 2003) and
Donoho reported that one of the benefits of a repro-
ducible computational workflow was improved team-
work (Donoho, 2010). Our experience would support
both of those ideas, as team members with variable
clinical, statistical, and technical backgrounds have
all contributed to the development of this workflow
and to the quality of the workflow’s research prod-
ucts.
In conclusion, reproducible reporting is a key
component of the reproducible research paradigm.
This manuscript presents an inexpensive, practical,
and easily adopted workflow for collaborative repro-
ducible biomedical reporting when working with sen-
sitive data.
ACKNOWLEDGEMENTS
We thank Dr. Michael Kahn for his comments and
suggestions on our manuscript. Dr. Bennett is sup-
ported by Eunice Kennedy Shriver National Insti-
tute for Child Health and Human Development Grant
K23HD074620.
REFERENCES
Chacon, S. and Straub, B. (2014). Pro git. Apress. Online
at https://git-scm.com/book/en/v2.
De Alwis, B. and Sillito, J. (2009). Why are software
projects moving from centralized to decentralized ver-
sion control systems? In Cooperative and Human
Aspects on Software Engineering, 2009. CHASE’09.
ICSE Workshop on, pages 36–39. IEEE.
Donoho, D. L. (2010). An invitation to reproducible com-
putational research. Biostatistics, 11(3):385–388.
Gandrud, C. (2015). Reproducible Research with R and
RStudio. Chapman & Hall/CRC Press, second edition.
Knuth, D. E. (1984). Literate programming. The Computer
Journal, 27(2):97–111.
Leisch, F. (2002). Sweave: Dynamic generation of statisti-
cal reports using literate data analysis. In H
¨
ardle, W.
and R
¨
onz, B., editors, Compstat 2002 — Proceedings
in Computational Statistics, pages 575–580. Physica
Verlag, Heidelberg. ISBN 3-7908-1517-9.
National Academies of Sciences, Engineering, and
Medicine, Division on Engineering and Physical Sci-
ences, Board on Mathematical Sciences and Their
Applications, Committee on Applied and Theoretical
Statistics (2016). Statistical Challenges in Assessing
and Fostering the Reproducibility of Scientific Results:
Summary of a Workshop. National Academies Press.
Peng, R. D. (2011). Reproducible research in computational
science. Science, 334(6060):1226–1227.
Peng, R. D., Dominici, F., and Zeger, S. L. (2006). Re-
producible epidemiologic research. Am J Epidemiol,
163(9):783–9.
Rossini, A. and Leisch, F. (2003). Literate statistical prac-
tice. Biostatistics Working Paper Series. Working Pa-
per 194. accessed May 17th, 2016.
Xie, Y. (2015). Dynamic Documents with R and knitr, Sec-
ond Edition. Chapman & Hall/CRC The R Series.
CRC Press.
Collaborative Reproducible Reporting - Git Submodules as a Data Security Solution
235
