
Semantic Knowledge Base Construction from Radiology Reports
Eriksson Monteiro, Pedro Sernadela, Sérgio Matos, Carlos Costa and José Luís Oliveira
Department of Electronics, Telecommunications and Informatics (DETI),
Institute of Electronics and Telematics Engineering of Aveiro (IEETA), University of Aveiro, Aveiro, Portugal
Keywords: Semantic Web, Healthcare Information Management, Clinical Reports, Radiology, Text-mining.
Abstract: The tremendous quantity of data stored daily in healthcare institutions demands the development of new
methods to summarize and reuse available information in clinical practice. In order to leverage modern
healthcare information systems, new strategies must be developed that address challenges such as extraction
of relevant information, data redundancy, and the lack of associations within the data. This article proposes
a pipeline to overcome these challenges in the context of medical imaging reports, by automatically
extracting and linking information, and summarizing natural language reports into an ontology model.
Using data from the Physionet MIMIC II database, we created a semantic knowledge base with more than
6.5 millions of triples obtained from a collection of 16,000 radiology reports.
1 INTRODUCTION
Nowadays, healthcare professionals recognize the
benefits of information technology (IT) for daily
clinical practice. During the last decades, researchers
have developed diverse solutions for improving
information storage, management and retrieval in
healthcare scenario (Thompson et al., 2014) (Belleau
et al., 2008). However, the tremendous
heterogeneous clinical data produced in distinct
healthcare centers is a critical issue (Howe et al.,
2008). Digital repositories containing clinical
information, assessment reports and guidelines are
usually available for consultation in those centers.
Still, these data are not always structured and
organized, hindering information retrieval and
knowledge extraction. Furthermore, even though it
is currently possible to find multiples sources of
medical information in healthcare centers, there is a
lack of integration between these data sources. For
example, traditional clinical information retrieval
systems are usually connected to a single type of
data source and do not enable information from
heterogeneous data sources to be connected and
queried. Additionally, online platforms such as
Radiopaedia (Gaillard and Jones, 2009), GoldMiner
(Kahn and Thao, 2007) and AuntMinnie (Minnie,
2002) provide rich collaborative repositories of
radiology cases and articles, but these systems do
not exploit linked data across similar online
platforms. Linking the information available on
these kinds of systems has great potential for
knowledge discovery in radiology.
One of the main issues that limit the
implementation of a solution that contemplates the
linked data scenario is related to how the
information is stored and provided. Typically, most
clinical information is commonly stored using
relational databases (e.g., Microsoft SQL Server,
MySQL) and queried through SQL (Structured
Query Language). According to Pathak et al. (Pathak
et al., 2012), relational model has several limitations
when compared to RDF (Resource Description
Framework) based solutions. Firstly, in terms of data
management process (i.e. add, update, delete), RDF
does not differentiate ontology classes and
properties from the instances of the ontology classes.
This makes it more flexible when compared to
relational models, which need to be reorganized if
database schema changes. Second, RDF resources
are identified by a globally unique URI, making it
possible to create references between two different
RDF graphs, even in completely different
namespaces, therefore enabling data linkage and
integration processes. Third, the relational model
does not have notion of hierarchy, which makes
difficult to apply SQL queries for reasoning
purposes. In opposition, these types of queries are
natively supported in RDF (RDF Schema) and OWL
(Web Ontology Language). Lastly, there is a lack of
Monteiro, E., Sernadela, P., Matos, S., Costa, C. and Oliveira, J.
Semantic Knowledge Base Construction from Radiology Reports.
DOI: 10.5220/0005709503450352
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 5: HEALTHINF, pages 345-352
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reser ved
345

a formal temporal model for representing relational
data. For instance, SQL provides minimal support
for temporal queries natively, in contrast to
SPARQL (SPARQL Protocol and RDF Query
Language) (Prud’Hommeaux and Seaborne, 2008)
that already provides these extensions. Concluding,
Semantic Web technologies provide an enhanced
model for making clinical and research data
available for secondary use and exploitation.
This article presents a complete pipeline that
comprises biomedical information extraction from
unstructured textual reports and the creation of a
knowledge base using Semantic Web and Linked
Data standards (Berners-Lee et al., 2001). Text-
mining techniques were used to extract relevant
information from clinical reports. Next, those
information elements were mapped to an adequate
ontology. The result is an enriched knowledge base
(in RDF format) from radiology reports. It aims to
support knowledge discovery processes and to serve
as a basis for the construction of decision support
systems.
This document is organized as follows: Section
II presents some related work, namely about the
application of text mining and Semantic Web in
biomedical and clinical systems. Section III
describes the proposed method to extract relevant
information from clinical reports and the resulting
semantic knowledge base structure. Section IV
discusses the results obtained with the achieved
pipeline. Finally, section V summarizes the
contributions and concludes the manuscript.
2 BACKGROUND
2.1 Text Mining & Clinical Text
Analysis
Handling and retrieving knowledge from biomedical
textual resources remains a challenge, mainly due to
the huge volume of data produced nowadays in
healthcare institutions. The development of text
mining algorithms and tools aims to support these
tasks. In the biomedical field, important text mining
contributions have focused on named entity
recognition (Campos et al., 2013a), which aims to
identify chunks of text associated to specific
biomedical entities of interest. Usually, it is a
complex task due to the domain specificity – large
set of terms, heterogeneous and ambiguous
concepts, dynamic terminology (Zhou et al., 2004).
Several tools, such as Whatizit (Rebholz-
Schuhmann et al., 2008), NCBO Annotator (Jonquet
et al., 2009), GIMLI (Campos et al., 2013a), Neji
(Campos et al., 2013b) and cTAKES (Savova et al.,
2010), apply machine learning and dictionary-based
methods, or a combination of these approaches to
solve this issue.
Some frameworks already provide services for
text analysis and knowledge extraction. For
example, UIMA (Ferrucci and Lally, 2004) and
GATE (Cunningham n.d.) are general frameworks
for developing complex information extraction
systems. These frameworks also provide enough
flexibility to build custom processing pipelines
based on software modules. Nevertheless, they are
too general and need to be tuned, or extended, for
improving their performance in specific domains.
Currently, it is already possible to find modules
optimized for the biomedical domain (e.g. JCoRe
(Hahn et al., 2008)), which are built on top of one of
these frameworks. There are also libraries such as
NLTK (Bird, 2006) and OpenNLP (Baldridge, 2005)
that provide several natural language processing,
machine-learning and text-mining methods. Finally,
tools such as Neji and cTAKES were specifically
developed for the biomedical domain, aiming to
provide a user-friendlier framework for building
text-mining solutions.
2.2 Semantic Web
Nowadays, the Semantic Web (SW) paradigm
(Berners-Lee et al., 2001) involves a broad set of
modern technologies that are used to link, exploit
and deliver knowledge for both machine and human
consumption.
Using state-of-the-art standards such
as RDF, OWL and SPARQL, SW can tackle
traditional data issues such as heterogeneity,
distribution and interoperability, providing an
interconnected network of knowledge. These
technologies emerged as a next-generation software
development paradigm and are appropriate for
dealing with the intrinsic interrelationships in the life
sciences field, providing improved computational
features to exchange and accurately interpret
knowledge.
Regarding the healthcare context, SW
technologies have been applied for transforming the
enormous quantity of data produced in useful
knowledge capable of improving clinical methods
and workflows. For instance, the development of
ontologies (Tao et al., 2011) and semantic
frameworks allowed answering time-oriented
queries through temporal relation inference in
clinical narrative reports (Tao et al., 2010). Another
study shows that SW inference and federated
HEALTHINF 2016 - 9th International Conference on Health Informatics
346
querying mechanisms can be used for cohort
identification from Electronic Health Records
(EHRs) (Pathak et al., 2012). Finally, other studies
demonstrate how SW can provide semantic
interoperability between disconnected clinical
domains (Laleci et al., 2013) or different healthcare
systems (Lopes and Oliveira, 2011), as well as
aiding and supporting clinical diagnosis through
well-structured ontologies (Bastiao Silva et al.,
2014). Healthcare systems are adopting SW for
building better solutions to represent and discover
knowledge contained in clinical data. However, its
integration with healthcare state-of-the-art systems is
not trivial. Current solutions are based on a set of
ETL (Extract-Transform-and-Load) techniques to
elevate the data to SW standards, requiring a
significant effort in data transformation and
ontology mapping processes. Regarding the
integration of text-mining results in SW, several
ETL procedures have been applied (Sernadela et al.,
2015) in order to translate information.
3 METHODS AND MATERIALS
3.1 Pipeline Overview
This article proposes a pipeline for creating a
knowledge base from radiology reports. The pipeline
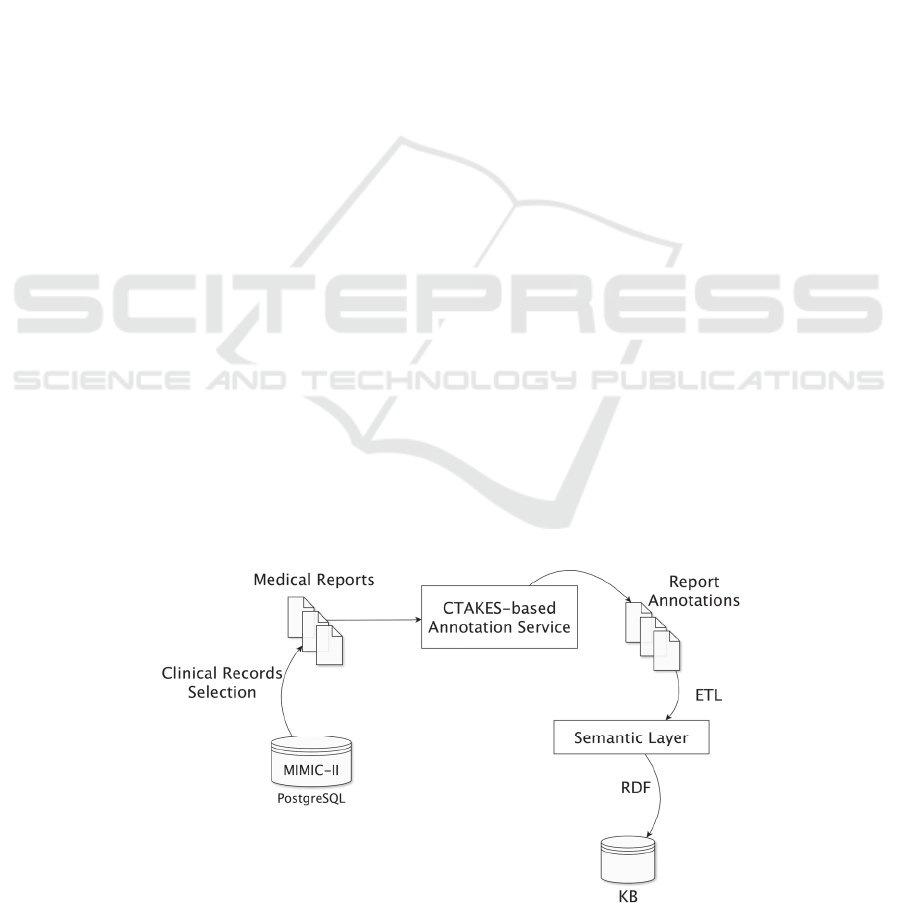
architecture is illustrated in the Fig. 1 and it consists
of five main blocks. In the first stage, clinical
records are selected and extracted from a public
database. Next, clinical free-text is obtained from
each record. In the third step, the free-text is
annotated through a dedicated service and the results
stored in separated objects. Later, a semantic layer
engine converts the annotations to the RDF format
using advanced ETL features. Finally, the resulting
RDF file is uploaded to a triple store database named
COEUS (Lopes and Oliveira, 2012), which allows
us to perform SPARQL queries for exploring the
information available in the knowledge base. The
pipeline modules and respective workflow will be
described in the next sections with more detail.
3.2 Clinical Reports Selection
The development and validation of our system was
performed using the radiology reports extracted
from the Multiparameter Intelligent Monitoring in
Intensive Care II (MIMIC-II) database. MIMIC-II is
a joint project of the MIT, Philips Medical System,
Philips Research North America and Beth Israel
Deaconess Medical Center. It aims to promote and
assess advanced patient monitoring systems (Saeed
et al., 2011). MIMIC-II is a PostgreSQL database
that contains data from more than 30,000 patients,
collected between 2001 and 2008. In our project, we
were interested in the information of 384,000
radiology reports. Moreover, we selected a subset
comprising approximately 16,000 of the latest
reports.
In data gathering process, it was necessary to
select and collect a subset of records that will
compose our case study. Next, the proposed pipeline
processed them, extracting information about
concepts and identifying respective relations for
building the knowledge base.
3.3 Annotation Service
The proposed pipeline contemplates a biomedical
clinical text annotation service that performs named-
entities recognition, concept recognition and relation
extraction (i.e. identifying relations between
Figure 1: Pipeline overview.
Semantic Knowledge Base Construction from Radiology Reports
347

concepts mentioned in the text). It was implemented
as a Representational State Transfer (REST) API
where the annotations are retrieved by making
HTTP POST requests to that service (Fig. 2).
Figure 2: This picture depicts the REST service for
annotating biomedical clinical free-text. The client sends
the texts using the HTTP POST method and receives the
annotation in the standoff format.
Scalability of the solution is ensured by
dynamically launched workers, named Pipeline
processors. The maximum number of workers used
to handle the annotation requests can be configured
when launching the service. There is also a
Processing Scheduler that manages the requests
distribution according with workers load, improving
the service throughput. This is a very important
issue, since the annotation process is relatively time
consuming. On average, it takes 1.5 seconds to
annotate each report in our dataset, when executed
on a virtual machine with 8 vCPU Intel(R) Xeon(R)
X5650 @ 2.67GHz and 8GB of RAM.
Regarding the format used to provide the
annotations, we decided to use a standoff format
similar to the one used in the BioNLP Shared Tasks
(Leech, 1993), where the annotations are stored
separately from the document text. It follows a
simple structure where each line contains one
annotation that has associated an assigned identifier
restricted to an entity (T), normalization (N) or
relation (R). An entity corresponds to a text-bound
annotation found on the plain-text report. If the
system can semantically classify the recognized
entity, it then associates a normalization annotation.
This corresponds to a semantic identifier of a given
database (e.g. Unified Medical Language System
(UMLS)). Additionally, a relation annotation can be
established if the system detects a relation between
two entities.
The system uses Apache cTAKES to implement
the entire clinical text processing. It is an open
source natural language processing tool for
extraction of information from clinical texts of
electronic medical records. Fig. 3 depicts the
pipeline implemented using several components of
the Apache cTAKES. Firstly, the document
processing stage includes segment detection,
sentences detection and tokenization, using the
OpenNLP Maximum Entropy package. In addition,
the SPECIALIST NLP Tools are used for dealing
with lexical variations in the clinical texts.
Moreover, to annotate syntactic structures and to
perform concept recognition, it was also used
specialized components provided by cTAKES,
namely annotators that combine rule-based with
machine-learning techniques. For example, the
cTAKES DictionaryLookup annotator used for
concept recognition tries to match spans of text to
dictionary entries.
In our case, it was used a dictionary built from
the 2014 UMLS Metathesaurus database. It contains
key terminology, classification and coding standards
assigned to terms. Each term has a concept unique
identifier (CUI) and an identifier for the semantic
type (TUI). The dictionary comprised terms from
five distinct semantic groups, each composed by a
Figure 3: cTAKES pipeline.
HEALTHINF 2016 - 9th International Conference on Health Informatics
348

set of semantic types (Table 1). The terms used to
define the medication semantic group were obtained
from the RXNORM database (Liu et al., 2005) while
the other terms for the remaining four semantic
groups where gathered from the SNOMEDCT
database (Stearns et al., 2001).
Table 1: Sets of semantic types used for defining the
semantic groups.
Semantic
GroupName
UMLSSemanticTypes
Medication
T073, T103, T109, T110, T111, T115,
T121, T122, T123, T130, T168, T192,
T195, T197, T200 and T203
Anatomical
site
T021, T022, T023, T024, T025, T026,
T029 and T030
Clinical
procedures
T059, T060 and T061
Clinical
disorders
T019, T020, T037, T046, T047, T048,
T049, T050, T190 and T191
Clinical
findings
T033, T034, T040, T041, T042, T043,
T044, T045, T046, T056, T057 and T184
In addition to concept recognition, the system
performs information extraction regarding clinical
site effects of drugs using the cTAKES
SiteEffectAnnotator. This component uses rule-based
methods for annotating site effect relations, allowing
us to recognize site effects and causative drugs in the
clinical texts.
Table 2: cTAKES annotators used for extracting binary
relation between identified concepts.
BinaryRelationAnnotator Description
DegreeOfRelation
ExtractorAnnotator
The component identifies
“degree of” relation between an
event and a modifier. As
example, degree of pain.
LocationOfRelation
ExtractorAnnotator
The component is used to
recognize the “location of”
relation between identified
concepts. For instance, location
of pain.
EventEvent
RelationAnnotator
The pipeline used this component
to annotate relation between two
consecutive event mentions. The
following are some event
samples: stable, change, evident
and process.
EventTime
RelationAnnotator
The component identifies
temporal relation between a time
mention and an event. For
example, for how long the patient
has been sick.
The extraction of binary relations between
concepts identified in clinical free-text is another
important feature for information retrieval systems
and was also exploited to enrich the quality of the
knowledge base resulting from our method. The
components used for binary annotation are described
in Table 2.
3.4 Semantic Layer
The model adopted for entity and normalization
employs domain ontologies and vocabularies,
creating extremely rich stores of metadata on Web
resources. The pipeline uses the AO (Annotation
Ontology) model to represent the clinical reports
annotations, producing enriched data with the
fragments of the annotated resource and respective
associated terms.
Fig. 4 shows an example of an annotation for the
word “chest” (i.e. the upper part of the trunk
between the neck and the abdomen) detected on a
medical report. The representation includes the
annotation URI (ao:Annotation), the clinical report
source (pav:SourceDocument) and the respective
data (ao:TextSelector). The model stores
information regarding the context of the annotation
(e.g. ann:T1_context) such as location of the
detected text in the report, the semantic group (e.g.
AnatomicalSiteMention), the semantic identifier (e.g.
C0817096) of the recognized text and the source
report identifier (e.g. 480). The ao:exact data
property denotes the entity recognized by the text-
mining tool. The concept normalization output is
defined through the ao:hasTopic property,
representing the semantic identifier of the annotated
entity. The ao:body property represents the entity
tag or domain detected by the text-mining tool.
An adaptable model that links and describes each
interaction is also used to establish relations between
them. Relation annotations are simple AO
annotations with the addition of two object
properties. The addiction of these properties allows
us to establish binary relations between entity
annotations and associated roles. Hence, we can
identify the source entity annotation through the
ann:argument property, and the respective target by
using the ann:target property. By using the ao:body
data property, we can establish the type of associated
relation. An overview of this model is represented in
the Fig. 5 example, where a unidirectional relation
between a “CT” (Computed Tomography) and the
“abdomen” (portion of the body that lies between
the thorax and the pelvis) is established. Basically,
it shows that a CT was performed in (location_of)
Semantic Knowledge Base Construction from Radiology Reports
349
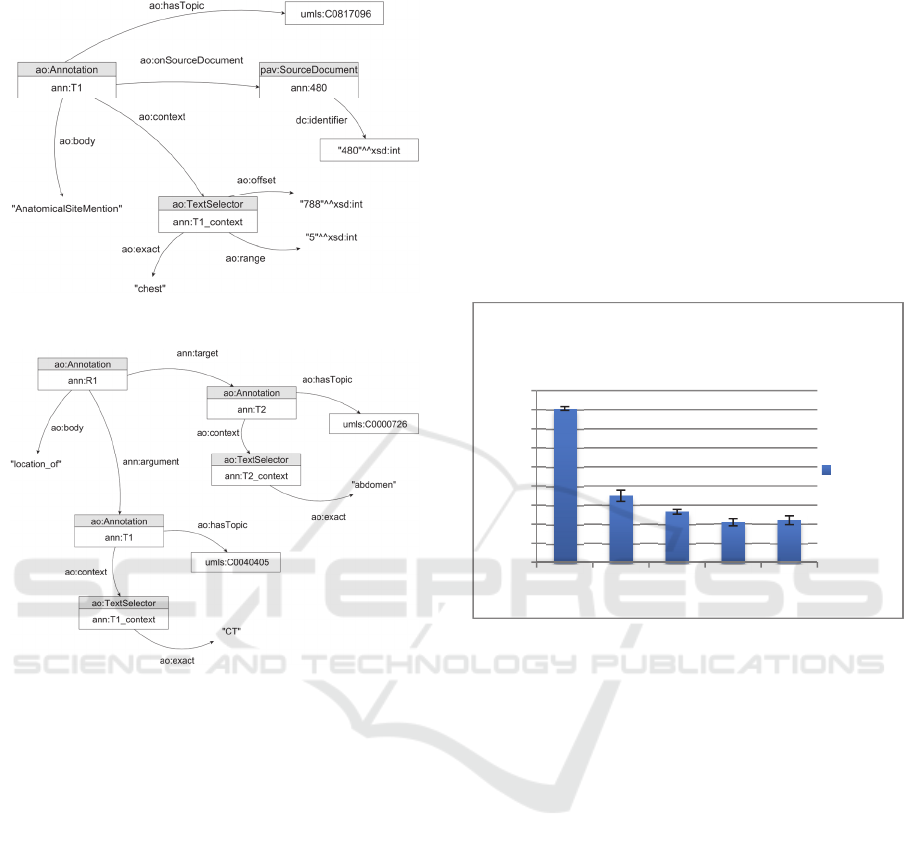
the abdomen.
Figure 4: Entity and normalization annotation model.
Figure 5: Relation annotation model.
4 RESULTS
The pipeline validation is achieved through a case
study that aimed to create a semantic repository
from a dataset of radiology clinical reports.
Moreover, it is expected to use the facts represented
in the knowledge base to serve in an inference
engine for reasoning. The dataset contained
approximately 16,000 clinical free-text documents
and our pipeline constructed an associated semantic
database with more than 6.5 million triples that were
stored in a triple store database using COEUS.
The REST service for annotating the biomedical
text is the most time consuming component of our
pipeline. So, during the development of the solution
we had this fact into consideration. The service uses
a processing scheduler that was implemented to take
advantage of multiprocessing. It is possible to define
several pipeline processors to handle the requests.
Each pipeline processor runs in a thread, which
allows us to take advantage of the multiple cores
available in the server. The impact of implementing
this kind of solution for our cTAKES-based
annotation service was evaluated. We tried several
set-up configurations, where we used different
number of launched pipeline processor. It was
measured the timespan to annotate 100 reports using
the annotation service. Furthermore, for each
experiment, we did 5 runs aiming for analyzing the
standard deviation in order to be more confident
with the results. The following picture shows the
annotation service performance while varying the
number of pipeline processors from 1 to 10
processors.
Figure 6: This picture shows how changing the number of
pipeline processors can improve the service performance.
By analyzing the results, we can observe that
using 8 processors we decreased the time needed to
process the reports by 74% compared with the
default usage of one cTAKES pipeline processor.
After processing the reports using our pipeline,
we were able to retrieve information stored in the
knowledge base using SPARQL queries
contemplating semantic identifiers and semantic
relations. Therefore, we could retrieve all distinct
reports where specific UMLS CUIs are present. We
were also able to exploit semantic properties
associated to relations established between concepts.
5 CONCLUSIONS
Clinical repositories represent one of the most
valuable resources for healthcare systems. They
contain relevant information for all clinical
stakeholders. However, most of the available clinical
repositories store patient reports in suboptimal ways,
hindering the application of knowledge discovery
0
50
100
150
200
250
300
350
400
450
1 P3 P6 P8 P10 P
Time for processing 100 reports (seconds)
Average
Time
HEALTHINF 2016 - 9th International Conference on Health Informatics
350

techniques. For this reason, our first contribution
was focused on the development of a complete text-
mining solution to extract meaningful information
from clinical narrative reports. This solution was
implement “as-a-service” using the cTAKES
framework. The main goal was to provide an easy
and functional service that can detect relevant
clinical concepts and their respective interactions.
As such, our developed tool can easily detect
entities, concepts and relations contained in clinical
text-reports. In addition, this work provides a
semantic knowledge base resulting from the
application of our method. This was built from the
clinical annotations retrieved from our text-mining
system. The constructed knowledge base was built
using Linked Data standards to facilitate the
application of several knowledge discovery
mechanisms, such as reasoning. Our final goal is to
make this radiology knowledge base freely available
through Physionet Web site
(http://www.physionet.org/). This will empower
novel discovery methods due to the existence of a
well-structured clinical report data.
ACKNOWLEDGEMENTS
This work has received support from the EU/EFPIA
Innovative Medicines Initiative Joint Undertaking
(EMIF grant n° 115372). Pedro Sernadela is funded
by Fundação para a Ciência e Tecnologia (FCT)
under the grant agreement SFRH/BD/52484/2014.
Eriksson Monteiro is funded by FCT under the grant
agreement SFRH/BD/102195/2014. Sérgio Matos is
funded under the FCT Investigator programme.
REFERENCES
Baldridge, J., 2005. The opennlp project. URL:
http://opennlp. apache. org/index. html,(accessed 2
February 2012).
Bastiao Silva, L., Costa, C. & Oliveira, J.L., 2014.
Semantic Search over DICOM Repositories. In
Healthcare Informatics (ICHI), 2014 IEEE
International Conference on. IEEE, pp. 238–246.
Belleau, F. et al., 2008. Bio2RDF: towards a mashup to
build bioinformatics knowledge systems. Journal of
biomedical informatics, 41(5), pp.706–16. Available
at: http://www.sciencedirect.com/science/article/pii/
S1532046408000415 [Accessed June 25, 2015].
Berners-Lee, T., Hendler, J. & Lassila, O., 2001. The
semantic web. Scientific American, 284.5, pp.28–37.
Available at: http://isel2918929391.googlecode.com/
svn-history/r347/trunk/RPC/Slides/p01_theSemantic
Web.pdf [Accessed July 8, 2014].
Bird, S., 2006. NLTK. In Proceedings of the
COLING/ACL on Interactive presentation sessions -.
Morristown, NJ, USA: Association for Computational
Linguistics, pp. 69–72. Available at:
http://dl.acm.org/citation.cfm?id=1225403.1225421
[Accessed June 25, 2015].
Campos, D., Matos, S. & Oliveira, J.L., 2013a. Gimli:
open source and high-performance biomedical name
recognition. BMC bioinformatics, 14(1), p.54.
Available at: http://www.biomedcentral.com/1471-
2105/14/54 [Accessed June 25, 2015].
Campos, D., Matos, S. & Oliveira, J.L., 2013b. Neji: a tool
for heterogeneous biomedical concept identification.
Proceedings of BioLINK SIG, 2013, pp.28–31.
Cunningham, H., GATE, a General Architecture for Text
Engineering. Computers and the Humanities, 36(2),
pp.223–254. Available at: http://link.springer.com/
article/10.1023/A%3A1014348124664 [Accessed June
25, 2015].
Ferrucci, D. & Lally, A., 2004. UIMA: an architectural
approach to unstructured information processing in the
corporate research environment. Natural Language
Engineering, 10(3-4), pp.327–348. Available at:
http://journals.cambridge.org/abstract_S13513249040
03523 [Accessed June 25, 2015].
Gaillard, F. & Jones, J., 2009. Collaborative Radiology
Resources: Radiopaedia. org as an Example of a Web
2.0 Radiology Resource. In AMERICAN JOURNAL
OF ROENTGENOLOGY. AMER ROENTGEN RAY
SOC 1891 PRESTON WHITE DR, SUBSCRIPTION
FULFILLMENT, RESTON, VA 22091 USA.
Hahn, U. et al., 2008. An overview of JCoRe, the JULIE
lab UIMA component repository. In Proceedings of
the LREC. pp. 1–7.
Howe, D. et al., 2008. Big data: The future of biocuration.
Nature, 455(7209), pp.47–50. Available at:
http://dx.doi.org/10.1038/455047a [Accessed January
28, 2015].
Jonquet, C. et al., 2009. NCBO annotator: semantic
annotation of biomedical data. In International
Semantic Web Conference.
Kahn, C.E. & Thao, C., 2007. GoldMiner: a radiology
image search engine. AJR. American journal of
roentgenology, 188(6), pp.1475–8. Available at:
http://www.ajronline.org/doi/full/10.2214/AJR.06.174
0 [Accessed June 25, 2015].
Laleci, G.B., Yuksel, M. & Dogac, A., 2013. Providing
semantic interoperability between clinical care and
clinical research domains. IEEE journal of biomedical
and health informatics, 17(2), pp.356–69. Available
at: http://www.ncbi.nlm.nih.gov/pubmed/23008263
[Accessed May 22, 2015].
Leech, G., 1993. Corpus Annotation Schemes. Literary
and Linguistic Computing, 8(4), pp.275–281.
Available at: http://llc.oxfordjournals.org/content/
8/4/275.short [Accessed June 25, 2015].
Liu, S. et al., 2005. RxNorm: prescription for electronic
drug information exchange. IT Professional, 7(5),
pp.17–23. Available at: http://ieeexplore.ieee.org/
Semantic Knowledge Base Construction from Radiology Reports
351

articleDetails.jsp?arnumber=1516084 [Accessed June
25, 2015].
Lopes, P. & Oliveira, J.L., 2011. A semantic web
application framework for health systems
interoperability. In Proceedings of the first
international workshop on Managing interoperability
and complexity in health systems - MIXHS ’11. New
York, New York, USA: ACM Press, p. 87. Available
at: http://dl.acm.org/citation.cfm?id=2064747.2064768
[Accessed April 23, 2013].
Lopes, P. & Oliveira, J.L., 2012. COEUS: “semantic web
in a box” for biomedical applications. Journal of
biomedical semantics, 3(1), p.11. Available at:
http://www.jbiomedsem.com/content/3/1/11
[Accessed March 5, 2013].
Minnie, A., 2002. AuntMinnie. com Launches New
Resource Focused on Diagnostic Imaging Centers.
Tucson, Arizona.
Pathak, J., Kiefer, R. & Chute, C., 2012. Using semantic
web technologies for cohort identification from
electronic health records for clinical research. AMIA
Summits on Translational Science Proceedings 2012.
Available at:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC33920
57/ [Accessed May 22, 2015].
Prud’Hommeaux, E. & Seaborne, A., 2008. SPARQL
query language for RDF. W3C recommendation, 15.
Rebholz-Schuhmann, D. et al., 2008. Text processing
through Web services: calling Whatizit.
Bioinformatics (Oxford, England), 24(2), pp.296–8.
Available at:
http://bioinformatics.oxfordjournals.org/content/24/2/2
96.short [Accessed June 25, 2015].
Rodríguez-González, A. et al., 2012. SeDeLo: using
semantics and description logics to support aided
clinical diagnosis. Journal of medical systems, 36(4),
pp.2471–81. Available at: http://www.ncbi.nlm.nih.
gov/pubmed/21537850 [Accessed April 10, 2015].
Saeed, M. et al., 2011. Multiparameter Intelligent
Monitoring in Intensive Care II: a public-access
intensive care unit database. Critical care medicine,
39(5), pp.952–60. Available at: http://www.
pubmedcentral.nih.gov/articlerender.fcgi?artid=31243
12&tool=pmcentrez&rendertype=abstract [Accessed
June 9, 2015].
Savova, G.K. et al., 2010. Mayo clinical Text Analysis
and Knowledge Extraction System (cTAKES):
architecture, component evaluation and applications.
Journal of the American Medical Informatics
Association : JAMIA, 17(5), pp.507–13. Available at:
http://jamia.oxfordjournals.org/content/17/5/507.abstra
ct [Accessed November 24, 2014].
Sernadela, P., Lopes, P. & Campos, D., 2015. A Semantic
Layer for Unifying and Exploring Biomedical
Document Curation Results. Bioinformatics and
Biomedical Engineering. Springer International
Publishing. Available at: http://link.springer.com/
chapter/10.1007/978-3-319-16483-0_2 [Accessed June
24, 2015].
Stearns, M.Q. et al., 2001. SNOMED clinical terms:
overview of the development process and project
status.
Proceedings / AMIA ... Annual Symposium.
AMIA Symposium, pp.662–6. Available at:
http://www.pubmedcentral.nih.gov/articlerender.fcgi?a
rtid=2243297&tool=pmcentrez&rendertype=abstract
[Accessed June 25, 2015].
Tao, C., Solbrig, H. & Chute, C., 2011. CNTRO 2.0: a
harmonized semantic web ontology for temporal
relation inferencing in clinical narratives. AMIA
Summits on Translational Science Proceedings 2011.
Available at: http://www.ncbi.nlm.nih.gov/pmc/
articles/PMC3248753/ [Accessed May 22, 2015].
Tao, C., Solbrig, H. & Sharma, D., 2010. Time-oriented
question answering from clinical narratives using
semantic-web techniques. The Semantic Web–ISWC
2010. Available at: http://link.springer.com/
chapter/10.1007/978-3-642-17749-1_16 [Accessed
May 22, 2015].
Thompson, R. et al., 2014. RD-Connect: an integrated
platform connecting databases, registries, biobanks
and clinical bioinformatics for rare disease research.
Journal of general internal medicine, 29 Suppl 3,
pp.S780–7.
Zhou, G. et al., 2004. Recognizing names in biomedical
texts: a machine learning approach. Bioinformatics
(Oxford, England), 20(7), pp.1178–90. Available at:
http://bioinformatics.oxfordjournals.org/content/20/7/1
178.short [Accessed May 28, 2015].
HEALTHINF 2016 - 9th International Conference on Health Informatics
352
