
Learning T2D Evolving Complexity from EMR and Administrative
Data by Means of Continuous Time Bayesian Networks
Simone Marini, Arianna Dagliati, Lucia Sacchi
and Riccardo Bellazzi
Biomedical Informatics Labs “Mario Stefanelli”, Dept. of Electrical, Computer and Biomedical Engineering,
University of Pavia, Pavia, Italy
Keywords: Type 2 Diabetes, Continuous Time Bayesian Network, Cohort Modeling, Disease Complexity.
Abstract: Predicting the complexity level (i.e. the number of complications and their related hospitalizations) in a T2D
cohort is a critical step in prevention, resource optimization and overall patient management. Our data set
was obtained by monitoring a T2D diabetic cohort along up to 10 years through electronic medical records
of a local healthcare agency data warehouse. In order to conveniently handle temporarily sparse data, we
designed a model describing the cohort evolution with Continuous Time Bayesian Networks (CTBN). The
network structure and its parameters are entirely data driven. Compared to traditional Bayesian Networks,
CTBNs admit cycles. As consequence, CTBNs fit the complexity of chronic metabolic syndromes where
variables show a reciprocal influence. Network nodes represent metabolic (glycated hemoglobin, lipid
profile (cholesterol, triglycerides), and biometric (BMI) data. We observed how these variables directly or
indirectly affect the disease level of complexity, and how the variables influence the cumulative adverse
events a patient undergoes.
1 INTRODUCTION
The development of computational tools to support
decisions in Medicine is driven by the increasing
burden of health care costs. Simulation models are
viable tools to predict health outcomes in target
populations and thus help optimizing resources
(Tarride et al., 2010; Marini et al., 2015). Diabetes is
a disease affecting a growing number of patients,
387 million individuals in 2014 with up to 592
million patients estimated by 2035 (International
Diabetes Federation, 2014; McEwan et al., 2014).
Providing tools to help clinicians assessing the
evolution of a diabetic cohort over time can
therefore help manage the high costs of diabetes.
The EU funded Mosaic project is dedicated to
the development of several models able to depict
meaningful clinical pathways in Type 2 Diabetes
(T2D) patients, in order to understand which clinical
and exogenous factors trigger worsened patients'
profiles. The dataset collected in a first project phase
is made up of 1020 T2D patients. These retrieved
information include data from hospital electronic
medical records (EMR) merged with administrative
information coming from Local Health Care Agency
(Dagliati et al 2014a). This means that observations
about patients are retrieved from follow-ups, which
usually occur every 6 month near the hospital where
subjects are in treated, and depict how metabolic
control and lipid profile evolve during the 5 to 10
years observation period. Clinicians record the
arising of complications due to the chronic diseases.
A complication is tied to its detection through a
set of prescribed exams and not to its physiological
onset specifically. This fact shows the importance of
the jointly analysis of clinical interventions and
physiological processes. To reach a comprehensive
understanding of the evolution of the disease we
leverage on the data coming from Local Health Care
Agency that include hospitalizations, drug purchases
and ambulatory encounters. Merging those sources
of data represents the first step to assess the disease
progression from both health care and management
points of view.
The exploitation of data mining methods allows
to automatically detect and reconstruct the most
frequent clinical temporal pathways patients
underwent (Dagliati et al. 2014b).
Since patient data streams are scattered along a
ten-year timeline, we decided to apply Continuous
Time Bayesian Networks (CTBN) (Nodelman and
al. 2002a) to design a network modelling TD2
338
Marini, S., Dagliati, A., Sacchi, L. and Bellazzi, R.
Learning T2D Evolving Complexity from EMR and Administrative Data by Means of Continuous Time Bayesian Networks.
DOI: 10.5220/0005708103380344
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 5: HEALTHINF, pages 338-344
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

trajectories. CTBNs have been successfully
exploited in Medicine (Gatti et al., 2012, Wang et
al., 2014) and Bioinformatics (Acerbi and Stella
2014, Liu et al., 2009).
2 METHODS
2.1 Variable Discretization
Firstly we have selected from the Mosaic data base a
set of meaningful clinical variables able to
characterize multiple aspects in the patients’ cohort:
metabolic control (hba1c), lipid profile (cholesterol
and triglycerides) and weight changes (BMI). All the
listed variables are relevant in T2D treatment and
control (American Diabetes Association, 2013;
Solano et al., 2006).
We defined the possible node states through
physiological parameters discretization. Discrete
states start from 0, with increasing values indicating
a progressive worsening of the patient condition. To
discretize variables, we used fixed threshold defined
by the Italian clinical guidelines for T2D care
(http://www.aemmedi.it/). Table 1 shows the states
and discretization thresholds of clinical variables.
Table 1: Number of states and discretization thresholds of
clinical variables.
Node’s state 0 1 2
HBA1c (mmol/mol) <59 ≥59 -
BMI <20 [20, 30] >30
CHOLESTEROL
(mg/dl)
<220
[220,
280]
≥280
TRIGLYCERIDES
(mg/dl)
<170
[170,
350]
≥350
Complications are tracked by worsening patient
condition and health care services accesses. Most of
the cost associated with diabetes is related to the
management of these complications (McEvan et al.,
2014). To assess the patient care complexity over
time, we defined a set of four complexity stages
(LOC, Level of Complexity) determined by the
number of complications and hospitalizations a
patient undergoes. Higher complexity means a
higher need for resources and a higher cost to
manage him/her (increased hospitalization rate,
more need for specific examinations, etc.). LOC is
an analytic indication of disease complexity
allowing to summarize the overall patient condition
in single number. We have defined complexity
levels and related status of network nodes as
follows.
STATUS 0: Stable patients. This segment belongs to
pre-diabetic patients and T2D patients not suffering
any complication yet.
STATUS 1: 1st level of complexity. Patients who are
starting to develop the first complication and require
punctual treatments. This stage and the following
one are built upon clinicians notes in the hospital
Electronic Medical Record (EMR). Complications
include Macro vascular event (i.e. Acute Myocardial
Infarction, Angina, Chronic ischemic heart disease,
Occlusion and stenosis of carotid artery, Peripheral
vascular disease, Stroke), Micro vascular event (i.e.
Diabetic Foot, Nephropathy, Retinopathy) and Not
Vascular event (i.e. Neuropathy, Fatty Liver
Disease).
STATUS 2: 2nd level of complexity. Patients with
multiple complications that need to be followed by
more than one specialist on a frequent basis.
STATUS 3: 3rd level of complexity. Patients likely
to suffer hospitalizations either due to the status of
their diabetes-related complications or because of
their metabolic instability. This stage triggers when
a previously developed complication leads to a
hospitalization that is detected through the
administrative data stream. Associations between
complications and hospital accesses have been
settled thanks to the collaboration with clinicians
(e.g. a hospitalization where the principal diagnosis
has been recorded with an IC9 code indicating a
disease of the genitourinary system after the onset of
Nephropathy).
2.2 Data Pre-processing and Data Set
Building
A first issue that we considered while analysing data
coming from a hospital EMR together with
administrative information is that T2D patients do
not start to be followed at the hospital immediately
after diabetes diagnosis. This happens because this
type of chronic patients are initially in charge of
their general practitioners who manage their cure
until it is more suitable for them to be followed in a
centre specialized in diabetes treatment.
For this reasons, observations do not show an
initial common time stamp, and some variables are
measured more frequently than other. Moreover,
these timestamps may vary also because of changes
in the patient conditions. For each time point we
have a direct observation for one or two variables
(e.g. cholesterol and triglycerides are often measured
together), but we lack information about unobserved
ones. Consequently, temporally sparse data with
Learning T2D Evolving Complexity from EMR and Administrative Data by Means of Continuous Time Bayesian Networks
339

missing values need to be pre-processed in order to
create suitable samples to be analysed with CTBNs
(e.g., the state of a variable was propagated until the
next known/measured value).
2.3 CTBN Machinery
Temporal dynamics are represented explicitly in
CTBNs. The system state space evolution is
described through conditional intensity matrices
(CIM) (Nodelman et al., 2002a). The number of
CIMs for each node is equivalent to the possible
state combinations of its parent nodes. For example,
if a child node C
1
has a parent node P
1
, and P
1
has
two states, then node C
1
is described by two CIMs.
If a child node C
2
has two parent nodes P
2
and P
3
,
with two and three states respectively, then C
2
is
described by six CIMs. CIMs are utilized to simulate
the evolution of the network state. Each CIM is a
square matrix, with a row for each state of the
described node, according the following schema:
CIM(C|p)
q
1(p)
q
12(p)
… q
1m(p)
q
21(p)
-q
2(p)
… q
2m(p)
…
…
… …
q
m1(p)
q
m2(p)
… -q
m(p)
The schema represents a CIM for the node C,
where p is the parent node status and m is the
number of states of C. In the example above, it could
be for instance P
1
=0 and P
2
=2. The elements q
i
on
the principal diagonal are related to the transition
time. In particular, when a node switches to the i-th
state, we utilize -q
i
to compute the time the node will
take before switching again. The time a node
remains in its state is randomly draw from an
exponential distribution with parameter q
i
,
∗
∗
, t ≥ 0.
Elements q
ij
not belonging to the principal
diagonal are always > 0 and they are utilized to
compute the next node state. Once a transition
happens (i.e. the time sampled by the distribution
above is expired), the node switches to another state
with probability
.
Note that for each row with m elements,
∑
0.
For example, if the first row a 3×3 CIM is [20 15
5], then the modeled node has three states, and the
first row models the behavior of the node in the first
state; the node remains in the first state for a time
sampled by a distribution with parameter
20. Once the node switches, it randomly ends in
the second state with probability 15/20 = 0.75, and
to the third state with probability 5/20 = 0.25.
To implement or CTBN, we utilized R Package
for Continuous Time Bayesian Networks (Shelton et
al., 2010).
2.4 Network Learning
The problem of learning the structure of a CTBN
from a data set D can be tackled as the problem of
finding a structure G maximizing a Bayesian score
(Nodelman et al., 2002b):
score
G∶D
ln
|
ln
While this problem is NP-hard in traditional
Bayesian Networks, it has been shown (Nodelman et
al., 2002b) that in CTBN it is possible to optimize
the parent set for each variable of the CTBN
independently. In other words, we can explore the
parent space of each variable with a local search to
find the best score. Once the maximum number of
parents local search is fixed, the search is
polynomial with respect to the number of variables
and the size of the data set (Nodelman et al., 2002b).
The aforementioned Bayesian score can be
decomposed in a sum of local contribution (i.e.
family scores). Each local score assess the quality of
a given putative parent set of a single variable. In
this way, we can optimize the parent set of each
variable taken singularly. For a more detailed
explanation of this process, we address the reader to
works of Nodelman (Nodelman et al., 2002a;
Nodelman et al., 2002b).
Note that no parameters had to be tuned for the
learning phase.
3 RESULTS
3.1 Network Structure
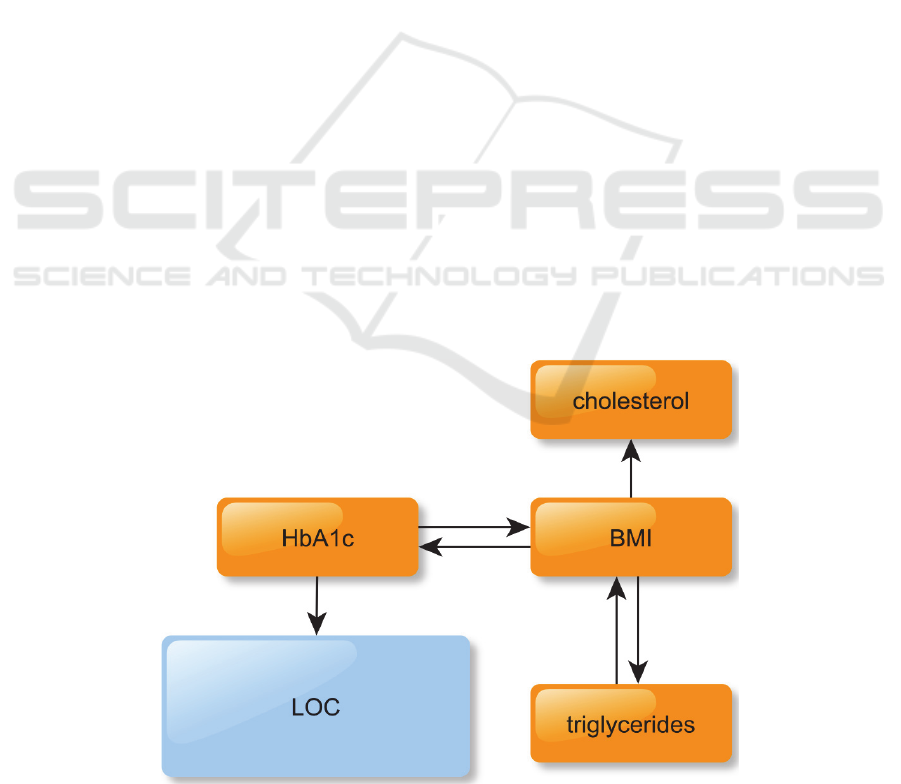
The resulting CTBN is shown in Figure 1. While the
only node directly affecting LOC is hbA1c, it is also
important to note that all the other nodes, with the
exception of cholesterol, are indirectly connected to
LOC through BMI and HbA1c. This means that the
model accounts for a wide variety of variables to
simulate the evolution of LOC. In other words, the
CTBN successfully learn meaningful variables
interconnections. HbA1c is a marker of long term
blood glucose concentration control, thus having a
pivotal role in diabetes monitoring.
Our network mimics how short-term changing
HEALTHINF 2016 - 9th International Conference on Health Informatics
340

variables (BMI influencing cholesterol and
triglycerides, and being influenced by triglycerides)
on the long run may raise the HbA1c level, which is
the key factor in determining the LOC and thus the
patient general health. In our learned network,
cholesterol does not influence LOC or any other
node. This does not mean cholesterol do not play a
role in the diabetes machinery, but rather than its
role does not emerge from our data.
3.2 Available Data Amount Influences
the Learned Network Structure
We assessed how the amount of available data
affects the network learning. We iteratively removed
an increasing number of random patient samples and
compared the learned network structure to the one
obtained from the full data set. In particular, we
gradually reduced our dataset by randomly removing
5% of the patients, and eventually reaching 50% of
the original data after 10 steps. We repeated 100
times this 10 step procedure, and we measured at
each step (a) if the network structure matches the
one learned on the full data set; and (b) the number
of the network edges of the newly learned network.
Results are shown in Figures 2 and 3.
Considering Figure 2, the amount of data seems
to be critical for this network design only when the
number of patients is less than 85% (~800 patients).
This means the very same network structure
emerges from data even if we remove 15% of the
available patients from our study (more than 95% of
the newly learned networks overlap the original).
This suggests the learned structure describes T2D
diabetes trajectories in a general and robust way.
However, this overlapping structure percentage
quickly drops once we utilize 75% or less of the
available patients. Similarly, as shown in Figure 3,
the edge number is reduced when fewer patients are
utilized for learning, leading to less rich and
interconnected networks.
3.3 Simulating LOC
We run a ten years long simulation with our
network. In particular, for each real patient, ten
artificial patients were simulated. Keeping a one-day
pace, the percentage of real (Rp) and simulated
patients (Sp) in a given LOC state were measured. It
was thus possible to calculate a daily LOC per-state
error as
|
|
, where
|
|
is the absolute value of
. The errors are depicted in Figure 4 with the blue
line. In general, we observed that the error grows
over time, but it is mostly below 15%. In fact, it is
always <10% for LOC 0, and it grows over 20%
only for LOC 1 after about three thousand days (thus
more than eight years after the beginning of the
simulation). Note that the granularity of the
simulation depends on the available data. In fact, in
CTBN we can simulate a step as short as the shortest
time unit utilized to measure our data. This means
that, since our EMR reported the date the patients
were visited and their data were recorded, we could
run simulations with a one day pace.
In order to validate our analysis, we proceeded
by randomly splitting our data into a training (85%)
and a test set (15%). We then re-learned the network
as from the training set only (the structure was
unaltered) and simulated the evolution of the learned
network for 10 years. The errors are measured as
explained above and depicted in Figure 4 with the
grey line. As expected, the errors on the test set were
higher, but still in a reasonable range, and below
25%.
4 CONCLUSIONS
In this work we designed a CTBN to describe a T2D
cohort. CTBN learning produced a meaningful
network fitting medical literature. Simulations
confirm that even for a ten year long timespan, error
in LOC is kept reasonably low (below 25%).
Moreover, the network is stable and it emerges from
data even if we remove 15% of the patient samples.
Although we obtained clinically meaningful
results, we are aware of some possible weakness in
this current approach. These weak aspects are
mainly related to the pre-processing of variable, like
Age or Time from Diagnosis, which naturally
increase during time. We are currently enhancing the
algorithm in order to make it able to process these
kinds of variables, taking into account that their
probability to change over time does not depend on
exogenous factors but it is intrinsic.
Furthermore, in future works, we aim to expand
our approach by including more network variables.
In particular, we will integrate EMR data with
administrative data about drug purchases to study
how different type of drugs (like Diabetes Therapies,
Antihypertensive and Statins) might cast an
influence on LOC.
REFERENCES
Acerbi, E., Stella, F. 2014. Continuous Time Bayesian
Networks for Gene Network Reconstruction: A
Learning T2D Evolving Complexity from EMR and Administrative Data by Means of Continuous Time Bayesian Networks
341
Comparative Study on Time Course Data.
Bioinformatics Research and Applications. Springer
International Publishing, 176-187.
American Diabetes Association 2013. Standards of
medical care in diabetes. Diabetes Care 36 (1): S11-
S66.
Dagliati, A., Sacchi, L., Bucalo, M., Segagni, D.,
Zarkogianni, K., Martinez Millana, A., Cancela, J.,
Sambo, F., Fico, G., Meneu Barreira, M.T., Cerra, C.,
Nikita, K., Cobelli, C., Chiovato, L., Arredondo, M.T.,
Bellazzi, R. 2014a. A Data Gathering Framework to
Collect Type 2 Diabetes Patients Data. Biomedical
and Health Informatics (BHI), 2014 IEEE-EMBS
International Conference Proceedings. 244 – 247.
Dagliati A., Sacchi, L., Cerra, C., Leporati, P., De Cata, P.,
Chiovato, L., Holmes, J.H., Bellazzi, R. 2014b.
Temporal Data Mining and Process Mining
Techniques to Identify Cardiovascular Risk-
Associated Clinical Pathways in Type 2 Diabetes
Patients Biomedical and Health Informatics (BHI),
2014 IEEE-EMBS International Conference
Proceedings. 240 – 243.
Gatti, E., Luciani, D., Stella, F. 2012. A continuous time
Bayesian network model for cardiogenic heart failure.
Flexible Services and Manufacturing Journal 4(4),
496—515.
International Diabetes Federation. 2014. IDF Diabetes
Atlas. 6th edn, 2014 Update. Brussels, Belgium:
International Diabetes Federation.
Liu, B., Thiagarajan, P.S., Hsu, D. 2009. Probabilistic
Approximations of Signaling Pathway Dynamics.
Computational Methods in Systems Biology. Springer
Berlin Heidelberg.
McEwan, P., Foos, V., Palmer, J.L., Lamotte, M., Lloyd,
A., Grant, D. 20014. Validation of the IMS CORE
Diabetes Model. Value Health (6):714-24.
Marini, S., Trifoglio, E., Barbarini, N., Sambo, F., Di
Camillo, B., Malovini, A., Manfrini, M., Cobelli, C.,
Bellazzi, R. 2015. A Dynamic Bayesian Network
model for long-term simulation of clinical
complications in type 1 diabetes. Journal of
Biomedical Informatics, ePub ahead of print.
Nodelman, U., Shelton, C.R., Koller, D. 2002a.
Continuous time bayesian networks. UAI02
Proceedings, 378—387.
Nodelman, U., Shelton, C.R., and Koller, D. 2002b.
Learning continuous time bayesian networks. In Proc.
of the 19th Conf. on Uncertainty in Artificial
Intelligence, pages 451–458.
Shelton, C.R., Fan, Y., Lam, W., Lee, J., Xu. J. 2010.
Continuous Time Bayesian Network Reasoning and
Learning Engine. The Journal of Machine Learning
Research 11: 1137-1140.
Solano, M.P., Goldberg, R.B. 2006. Lipid management in
type 2 diabetes. Clinical Diabetes 24(1): 27-32.
Tarride, J.E., Hopkins, R., Blackhouse, G., Bowen, J.M.,
Bischof, M., Von Key-serlingk, C., O’Reilly, D., Xie,
F., Goeree, R. 2010. A review of methods used in long-
term cost-effectiveness models of diabetes mellitus
treatment. Pharmacoeconomics. 28(4):255-77.
Wang, X., Sontag, D., Wang, F. 2014. Unsupervised
Learning of Disease Progression Models. Proceedings
of the 20th ACM SIGKDD international conference
on knowledge discovery and data mining, 85-94.
APPENDIX
Figure 1: The learned CTBN network.
HEALTHINF 2016 - 9th International Conference on Health Informatics
342
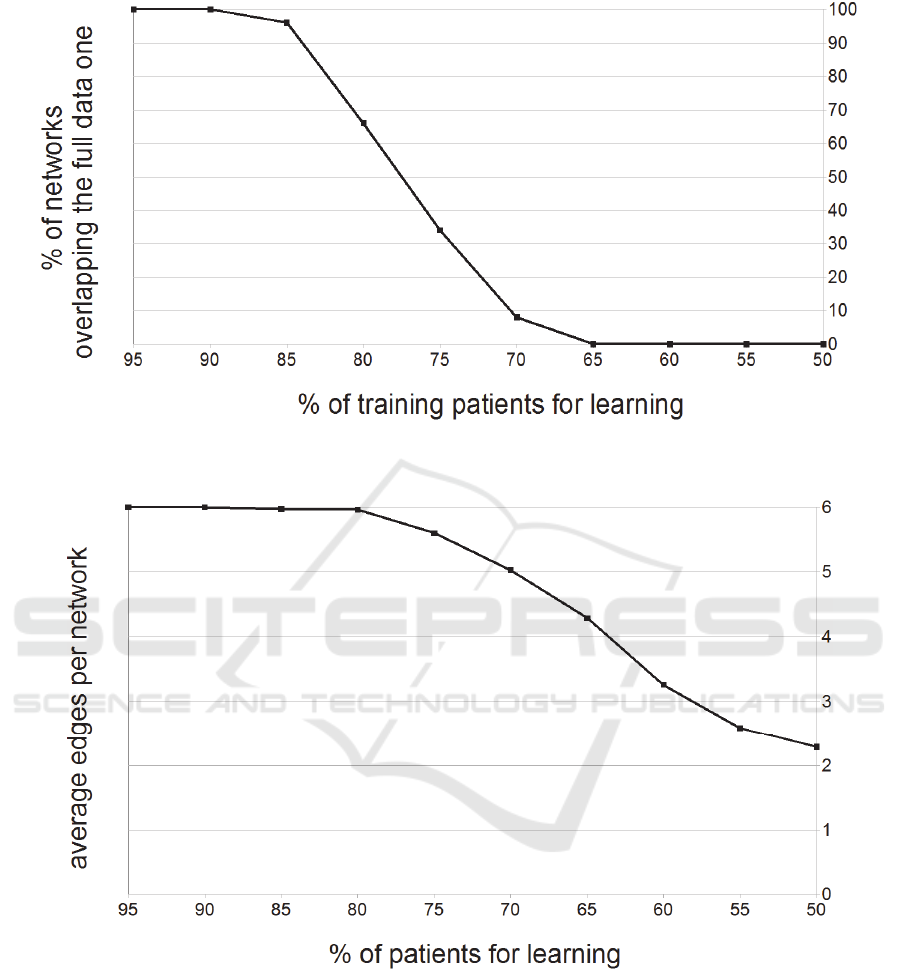
Figure 2: Percentage of newly learned network overlapping with the original one depends on the number of patients
sampled from the original data set.
Figure 3: Average edges per learned network depends on the fraction of patients sampled from the original data set.
Learning T2D Evolving Complexity from EMR and Administrative Data by Means of Continuous Time Bayesian Networks
343

Figure 4: Error for each state of LOC node. Error is measured as the absolute difference between the amount of real and
simulated patients in a given state, at a given time (one day pace). The error utilizing all data is represented in blue, while
the error on a test set (not utilized for training) is in grey.
HEALTHINF 2016 - 9th International Conference on Health Informatics
344
