
microRNA Detection with an Active Nanodevice F
o
F
1
-ATPase
Yao-Gen Shu and Zhong-Can Ou-Yang
Institute of Theoretical Physics, Chinese Academy of Sciences, Beijing 100190, China
Keywords:
Molecular Motor, F
o
F
1
-ATPase, Chromatophore, Processive Rotating, Ultrasensitive Biosensor.
Abstract:
A novel nanodevice was constituted with a rotary motor and a “battery”, F
o
F
1
-ATPase and chromatophore.
The former can processively rotate at about 10
3
r.p.m for more than one hour once the latter was recharged
by shine. If the nanodevice is captured by a target such as miRNA and processively rotates for 30 minutes,
the number of targets will be amplified by 10
5
ATP molecules. The sensitivity of detection was lower than
1.0 pM. Therefore, this method has potential to be developed into an ultrasensitive biosensor to detect low
expressed targets such as miRNA.
1 INTRODUCTION
F
o
F
1
-ATPase is the ubiquitous enzyme that uses the
transmembrane electrochemical potential to synthe-
size ATP in bacteria, chloroplasts and mitochondria.
The holoenzyme can be divided into two rotary mo-
tors, F
o
and F
1
. F
1
motor consists of a crown type
“stator” (α
3
β
3
) and a eccentric “rotor” (γ), while F
o
motor consists of a “stator” (a subunit) embedded in
membrane and a ring channels “rotor” (c
n
). The two
“stators” are fixed by b
2
and δ subunits, while the two
“rotors” are mechanically coupled by ε subunit. The
membrane embedded F
o
unit converts the proton mo-
tive force(p.m.f) into mechanical rotation of the “ro-
tor”, thereby causing cyclic conformational change of
α
3
β
3
crown (“stator”) in F
1
and driving three ATP
molecules synthesis for each rotation at nearly 100%
efficiency(Boyer, 1997; Noji et al., 1997; Yasuda
et al., 1998; Abrahams et al., 1994; Diez et al., 2004;
Toyabe and Muneyuki, 2015; Shu et al., 2010).
In vitro, however, F
o
F
1
-ATPase must be recon-
stituted in polymersome and coupled with Bacteri-
ochlorophyll to maintain its ATP synthesis, where
the Bacteriochlorophyll converted the light energy
into transmembrane p.m.f(Choi and Montemagno,
2005). The polymersome with Bacteriochlorophyll
was named chromatophore. The combination of
F
o
F
1
-ATPase and chromatophore is a sophisticated
nanomachine, in which chromatophore function as a
“battery” to power the rotary motor F
o
F
1
-ATPase, as
well as the ”battery” can remotely be recharged by
shine. Furthermore, the combination of F
o
F
1
-ATPase
and chromatophore can conveniently be prepared by
the phototrophic bacterium, in which the cells were
disrupted by sonication on ice. Each chromatophore
vesicle of 100 nm diameter contains on average one
F
o
F
1
-ATPase(Feniouk et al., 2002).
Additionally, molecular simulation(Shu and Lai,
2008) and nanoporous membrane experiment(Dong
et al., 2011) have demonstrated that the motor can
achieve about 10
3
r.p.m at saturated substrate concen-
tration and the chromatophore can processively power
the motor for more than one hour once the “battery”
was recharged by shine(Zhang et al., 2005; Deng
et al., 2007; Cheng et al., 2010), which means one
motor can generate about 10
5
ATP molecules during
30 minutes. On the other hand, microRNA(miRNA)
is often the marker of early diagnoses of cancer. Thus,
there is an urge to have a high sensitive detection of
miRNA due to its low expression in early cancer cell.
Here, we constituted a nanodevice with the combina-
tion of F
o
F
1
-ATPase and chromatophore to detect the
miR26a, a marker of hepatocarcinoma.
2 EXPERIMENTAL RESULTS
As Fig.1 shows, when target microRNA was base-
paired between capture probe and detection probe,
the motor embedded in chromatophore can be linked
to the magnetic beads surface, while the free motors
can be washed away as shown in Fig.2. The num-
ber of captured motors is equal to that of microR-
NAs because of there is only one ε subunit in F
o
F
1
-
ATPase motor. The amount of ATP generated by
F
o
F
1
-ATPase, thus, is in direct proportion to the num-
166
Shu, Y-G. and Ou-Yang, Z-C.
microRNA Detection with an Active Nanodevice F
o
F
1
-ATPase.
DOI: 10.5220/0005692301660169
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 1: BIODEVICES, pages 166-169
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
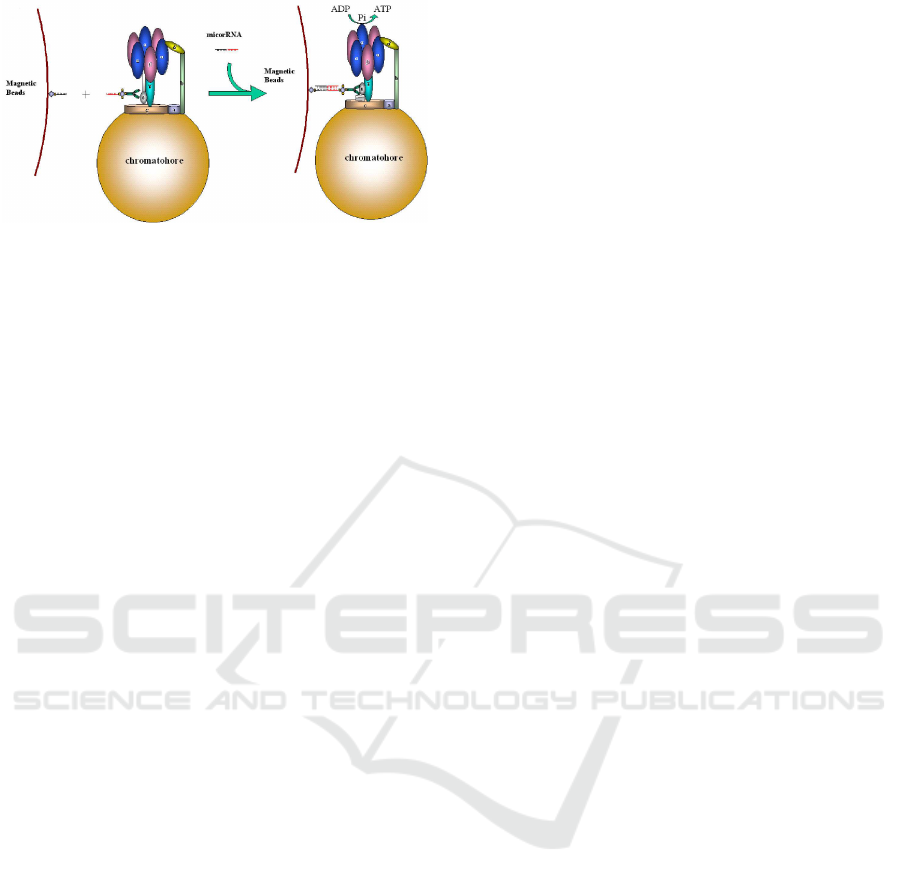
Figure 1: Cartoon of the biosensor based on ε subunit con-
jugation(not to scale). The hybridized motors are captured
by the magnetic beads, while the free motors will be washed
away. Thus, the number of the hybridized miRNA is pro-
portion to that of motors.
ber of motors and time of synthesis based on the same
storage energy in chromatophore. Here, the motor
function as an amplifier in which the number of target
microRNA is amplified to 10
5
times ATP molecules
during 30 minutes. The detector is able to be sensitive
to 1.0 pM of miR26a. Therefore, the active nanode-
vice has a potential to be developed into a dynamic
biosensor.
3 MATERIALS AND METHODS
3.1 Cell Lines and Reagents
Thermomicrobium roseum wa0073 (ATCC27502)
was purchased from ATCC (USA). The lu-
ciferase/luciferin ATP detection kits were pur-
chased from Promega Corporation (USA). ADP,
(+)-biotin N-hydroxysuccinimide ester and Neu-
trAvidin were purchased from Sigma-Aldrich (St.
Louis, USA). The microRNA extraction kits were
purchased from Tiangen, and the RNA probes
were synthesized by Sangon Biotech (Shanghai)
Co., Ltd.. The miR26a capture probe sequence
was 5’-biotin-AAAAAAAAAAAGCCTATCCT-
3’, the detection probe sequence was 5’-
GGATTACTTGAAAAAAAAAAAA-biotin-3’
and 2
′
Ome modified miR26a RNA sequence (UU-
CAAGUAAUCCAGGAUAGGCU) was synthesized
by Gene Pharma company (Shanghai). Streptavidin
magnetic beads (1.0 µm, Dynabeads
R
MyOneTM
streptavidinC1) was purchased from Life Technolo-
gies, Inc. The microplate luminometer was a Centro
XS3 LB 960 (Germany).
3.2 Preparation of Chromatophore
Containing the F
o
F
1
-ATPase
The chromatophore containing the F
o
F
1
-ATPase was
isolated and purified according to our previous pub-
lished protocols(Cheng et al., 2010; Shu and Ou-
Yang, 2012). Thermomicrobium roseum was cultured
at 60
◦
C for 24 h, and the cells were collected by cen-
trifugation at 4,000 rpm for 20 minutes. The pellets
were resuspended in 20 ml buffer A (pH 6.0), contain-
ing 20 mM Tricine-NaOH (pH 6.0), 2 mM MgCl
2
,
100 mM NaCl, and 10% glycerin (v/v), and sonicated
for 3 minutes. The lysate was centrifuged at 8000
rpm with R20A2 rotor for 30 minutes at 4
◦
C, and the
supernatant was collected and centrifuged at 40,000
rpm for 90 minutes at 4
◦
C. The precipitate contain-
ing the chromatophores was resuspended in buffer B
(pH 8.0), with 20 mM Tricine-NaOH (pH 8.0), 2 mM
MgCl
2
, 100 mM NaCl, and 10% glycerin (v/v), and
stored at −80
◦
C for further use.
3.3 Preparation of Monoclonal
Antibodies of ε-subunit
The ε-subunit was expressed and purified as in
Ref.(Su et al., 2006). The ε-subunit monoclonal
antibodies were prepared according to the method
for monoclonal antibody production procedure in
Ref.(Hanly et al., 1995), purified by precipitation with
33% (NH
4
)
2
SO
4
at 4
◦
C for 12 h, and the IgG parts
were separated using Sephadex G-200 and stored at
−20
◦
C before use.
3.4 Preparation of Detection
Probe-conjugated Motor
The motor with miR26a detection probe were pre-
pared by conjugating the streptavidin to the ε subunit
of F
o
F
1
-ATPase embedded in chromatophore through
biotinylated ε subunit monoclonal antibody(shown
in Fig.1). Then the biotinnylated miR26a detection
probe was bound to streptavidin. Specifically, 15 µl
of chromatophores (50 mg/ml) and 8 µl (0.5 mg/ml)
of biotinylated ε-subunit monoclonal antibodies were
mixed, diluted with 1 ml with PBS buffer, and then
incubated at 37
◦
C for 60 minutes. The free ε subunit
monoclonal antibodies were washed away by cen-
trifugation at 40,000 rpm for 20 minutes at 4
◦
C. The
precipitate was resuspended in 500 µl of PBS buffer.
Then, 7.5 µl (0.1 mg/ml) of NeutrAvidin was added
and diluted into 1 ml with PBS buffer, and incubated
at 37
◦
C for 10 minutes. The free neutravidin was then
washed away by centrifugation at 40,000 rpm for 20
microRNA Detection with an Active Nanodevice F
o
F
1
-ATPase
167
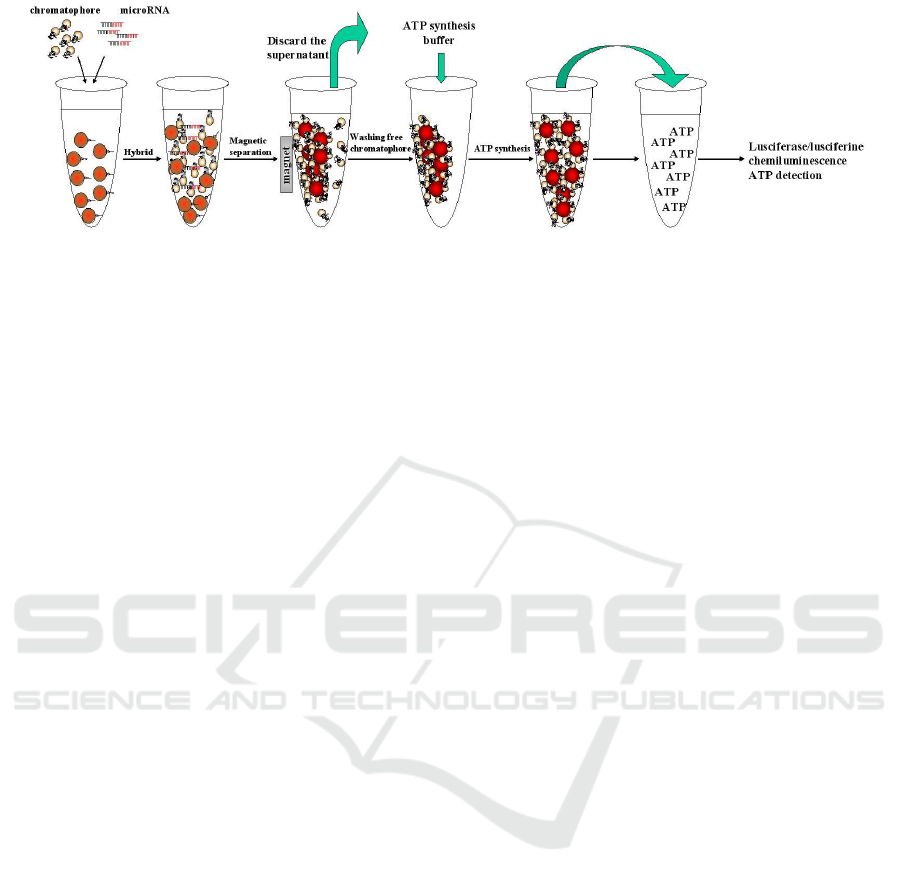
Figure 2: The procedure of miRNA detection. The first step is hybridization of targetmicroRNA; The second step is separation
between the captured motors and free ones. The free motors will be wash away, while the number of captured motors is equal
to that of microRNAs because of there is only one ε subunit in F
o
F
1
-ATPase motor; The third step is ATP synthesis in the
captured motors during 30 minutes; The final step is measure of ATP concentration with luciferase-luciferin.
minutes at 4
◦
C. The precipitate was resuspended in
500 µl of PBS buffer. Then, 10 µl (1µM) of biotiny-
lated detection probe were added, diluted into 1 ml
with PBS buffer, and incubated at 37
◦
C for 10 min-
utes. The free detection probes were then washed
away by centrifugation at 40,000 rpm for 20 minutes
at 4
◦
C. The precipitate was resuspended in 500 µl of
PBS buffer.
3.5 Preparation of Capture
Probe-conjugated Magnetic Beads
The magnetic beads conjugated with miR26a cap-
ture probe were prepared by conjugating the strep-
tavidin magnetic beads with the 5
′
-biotin modified
miR26a capture probe according to the manufac-
turer’s procedure(shown in Fig.1). Specifically, 100
µl 10 mg/ml streptavidin magnetic beads were pipet-
ted into a microtube and washed twice with 100 µl
1×B&W buffer (pH 7.5, 5 mM Tris-HCl, 0.5 mM
EDTA, 1 M NaCl). The magnetic beads were resus-
pended in 100 µl 2×B&W buffer. Then 100 µl bi-
otinylated capture probe in H
2
O was added and incu-
bated for 15 minutes at 25
◦
C using gentle rotation for
the binding reaction. After the binding reaction, the
conjugated magnetic beads were washed three times
with 1×B&W Buffer. In the end, the capture probe-
conjugated magnetic beads were resuspended in PBS
buffer and stored at 4
◦
C for usage.
3.6 miRNA Hybridization with Capture
Probe-conjugated Magnetic Beads
and Detection Probe-conjugated
Motor
30 µl of complementary miR26a (for sensitivity test)
in appropriate concentrations (a series of concentra-
tions ranging from 1 pM to 100 pM) was incubated
with 30 µl of 10 mg/ml magnetic beads with capture
probe in 20 µl hybridization buffer (final concentra-
tion is 5×SSC, 5× denhardt’s solution) at 37
◦
C for
30 minutes. Then the magnetic separation was per-
formed to removethe un-hybridized target microRNA
and resuspended the magnetic beads with 50 µl hy-
bridization buffer. For the binding of detection chro-
matophore, 30 µl detection probe-conjugated chro-
matophore was added into the magnetic beads and
incubated for 10 minutes at 37
◦
C. After the hy-
bridization was completed, the three times washing
with PBST and PBS were performed respectively to
remove the unbound chromatophore completely as
shown in Fig.2.
3.7 ATP Synthesis Assay
The ATP synthesis activity of F
o
F
1
-ATPase within the
chromatophores was determined using the luciferin-
luciferase method. The the magnetic beads were
resuspended with 100 µl ATP synthesis buffer (10
mM Tricine NaOH (pH 8.0), 5 mM MgCl
2
, 5 mM
Na
2
HPO
4
, 0.3 mM ADP 10% glycerol) and incu-
bated at 37
◦
C for 60 minutes, then separated by mag-
netic separator. 30 µl supernatant was transported
into the 96-well plate with three times, and 30 µl lu-
ciferase/luciferin working solution was added into, fi-
nally the chemiluminescence signals displayed in mi-
croplate luminometer were recorded immediately as
shown in Fig.2.
The relative light density emitted from luciferase-
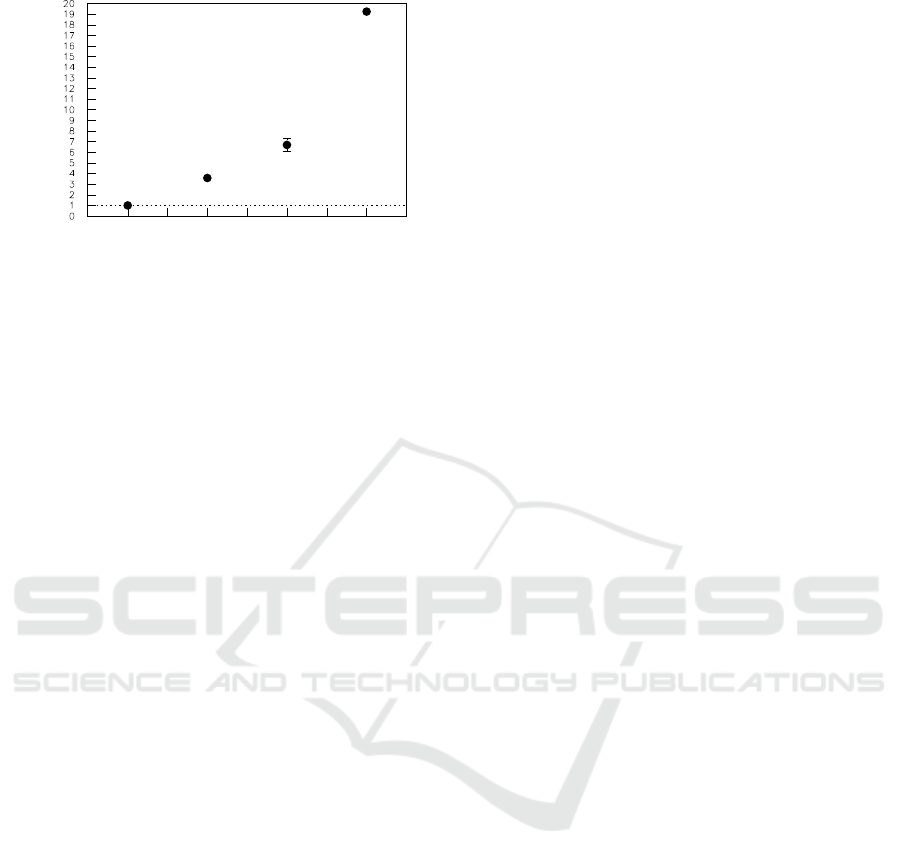
luciferin at different miR26a concentration is shown
in Fig.3, in which the light density of bufferis normal-
ized. The results indicated that the measured value
is in direct proportion to the miR26a concentration.
The results means that the sensitivity of detection was
lower than 1.0 pM.
BIODEVICES 2016 - 9th International Conference on Biomedical Electronics and Devices
168
0 1 10 100
miR26a (pM)
Relative Value
Figure 3: The relation between relative measured
value(mean± s.e.m.) and miR26a concentration. “0” cor-
responds to the buffer, and its light density is normalized.
Data points represent an average of 10-15 samples.
4 CONCLUSIONS
We have developed a novel nanodevice constituted
with a rotary motor and a “battery”, F
o
F
1
-ATPase and
chromatophore. The former can processively rotate at
about 10
3
r.p.m for more than one hour once the latter
was recharged by shine. If the nanodevice is captured
by a target such as miRNA and processively rotate for
30 minutes, the number of targets will be amplified by
10
5
ATP molecules. The sensitivity of the detection
was lower than 1.0 pM. This method has potential to
be developed into an ultrasensitive biosensor to detect
low expressed targets such as miRNA.
ACKNOWLEDGEMENTS
This work is supported by the National Basic Re-
search Program of China (973 Program) under grant
No. 2013CB932804 and the National Natural Science
Foundation of China under Grant No. 11574329 and
11322543.
REFERENCES
Abrahams, J. P., Leslie, A. G. W., Lutter, R., and Walker,
J. E. (1994). Structure at 2.8˚a resolution of F
1
-
ATPase from bovine heart mitochondria. Nature,
370:621–628.
Boyer, P. D. (1997). The ATP synthase-a splendid molecu-
lar machine. Annu. Rev. Biochem., 66:717–749.
Cheng, J., Zhang, X. A., Shu, Y. G., and Yue, J. C. (2010).
F
o
F
1
-ATPase activity regulated by external links on β
subunits. Biochem. Biophys. Res. Commun., 391:182–
186.
Choi, H. J. and Montemagno, C. D. (2005). Artificial or-
ganelle: Atp synthesis from cellular mimetic polymer-
somes. Nano. Lett., 355:2538–2542.
Deng, Z. T., Zhang, Y., Yue, J. C., Tang, F. Q., and Wei,
Q. (2007). Green and orange CdTe quantum dots as
effective pH-sensitive fluorescent probes for dual si-
multaneous and independent detection of viruses. J.
Phys. Chem. B, 111:12024–12031.
Diez, M., Zimmermann, B., B¨orsch, M., K¨onig, M.,
Schweinberger, E., Steigmiller, S., Reuter, R.,
Felekyan, S., Kudryavtsev, V., Seidel, C. A. M., and
Gr¨aber, P. (2004). Proton-powered subunit rotation
in single membrane-bound F
o
F
1
-ATP synthase. Nat.
Struct. Mol. Biol., 11:135–141.
Dong, H., Nie, R., Hou, X., Wang, P., Yue, J., and Jiang,
L. (2011). Assembly of F
o
F
1
-ATPase into solid state
nanoporous membrane. Chem. Commun., 47:3102–
3104.
Feniouk, B. A., Cherepanov, D. A., Voskoboynikova, N. E.,
Mulkidjanian, A. Y., and Junge, W. (2002). Chro-
matophore vesicles of Rhodobacter capsulatus con-
tain on average one F
o
F
1
-ATP synthase each. Bio-
phys. J., 82:1115–1122.
Hanly, W. C., Artwohl, J. E., and Bennett, B. T. (1995).
Review of polyclonal antibody production procedures
in mammals and poultry. ILAR J., 37:93–118.
Noji, H., Yasuda, R., Yoshida, M., and Kinosita, K. (1997).
Direct observation of the rotation of F
1
-ATPase. Na-
ture, 386:299–302.
Shu, Y. G. and Lai, P. Y. (2008). Systematic kinetics study
of F
o
F
1
-ATPase. J. Phys. Chem. B, 112:13453–13459.
Shu, Y. G. and Ou-Yang, Z. C. (2012). F
o
F
1
-
ATPase stator regulation studied with a reso-
nance model. In Proceedings of the International
Conferenceon Biomedical Electronics and Devices,
doi=10.5220/0003753401320137:132–137.
Shu, Y. G., Yue, J. C., and Ou-Yang, Z. C. (2010). F
o
F
1
-
ATPase, rotary motor and biosensor. Nanoscale,
2:1284–1293.
Su, T., Cui, Y., Zhang, X., Liu, X., Yue, J., Liu, N., and
Jiang, P. (2006). Constructing a novel nanodevice
powered by δ-free F
o
F
1
-ATPase. Biochem. Biophys.
Res. Commun., 350:1013–1018.
Toyabe, S. and Muneyuki, E. (2015). Single molecule ther-
modynamics of atp synthesis by f
1
-atpase. New J.
Phys., 17:015008.
Yasuda, Y., Noji, H., Kinosita, K., and Yoshida, M. (1998).
F
1
-ATPase is a highly efficient molecular motor that
rotates with discrete 120
◦
steps. Cell, 93:1117–1124.
Zhang, Y. H., Wang, J., Cui, Y., Yue, J., and Fang, X.
(2005). Rotary torque produced by proton motive
force in F
o
F
1
-ATP motor. Biochem. Biophys. Res.
Commun., 331:370–374.
microRNA Detection with an Active Nanodevice F
o
F
1
-ATPase
169
