
Temporally Synchronized Reversible Data Hiding of EEG to MREG
Angelos Fylakis
1, 2
, Anja Keskinarkaus
1, 2
, Vesa Kiviniemi
3, 2
and Tapio Seppänen
1, 2
1
Center for Machine Vision and Signal Analysis, University of Oulu, Oulu, Finland
2
MRC Oulu, Oulu University Hospital, Oulu, Finland
3
Oulu Functional Neuroimaging, MIPT Research Group, University of Oulu, Oulu, Finland
Keywords: Data Hiding, Data Management, Brain Research, Multimodality.
Abstract: Simultaneous MREG and EEG recordings are vastly used in neurobiology, but so far they are stored and
handled as separate files. This paper proposes a method to combine those two entities with the objective of
establishing data management efficiency, while secondary objectives are confidentiality, availability and
reliability in data. To be more specific, it is a reversible data hiding method for hiding EEG in MREG with
the ability of fully recovering MREG and the embedded EEG signal. It is based on histogram shifting,
exploiting data quantization and Region of Interest segmentation. The embedding procedure maintains
temporal synchronization between EEG and 32-bit MREG making it a novel data hiding application. It is
demonstrated through experiments that MREG maintains high perceptual fidelity and also verified that after
EEG extraction and acquisition of every electrode’s sample, MREG is fully reversed to its exact initial state.
1 INTRODUCTION
Modern ultrafast Magnetic Resonance Imaging
(MRI) sequences combined with multimodal data
produce vast amounts of data creating efficiency and
security related problems. Considering this together
with Human Connectome projects it is clear that
demands on data storage and analysis are constantly
increased. In such applications, data is stored in big
databases called electronic healthcare records. They
are either based on local hospital networks or cloud
networks. This paper presents a method to increase
data management efficiency and data security, which
are the main problems in most e-health applications.
The proposed method applies data hiding
techniques and embeds Electroencephalography
(EEG) data in MR-Encephalography (MREG)
recordings. MREG sequences are similar to fMRI
enabling even faster and more sensitive monitoring
of functional activation of the brain sampled every
25-100 msec. EEG-fMRI/MREG signal recordings
appear to have great importance in neurobiology
enabling researchers to understand neural behaviour.
fMRI/MREG provides detailed spatial resolution
showing activated brain areas but not details as to
temporal resolution. EEG, on the other hand,
provides information related to temporal resolution
promoting study on the dynamics of brain function,
while its poor spatial resolution restricts
identification of the neural sources (Menon and
Crottaz-Herbette, 2005). Overall, this is what makes
EEG and fMRI/MREG complementary data.
The proposed method will focus on the
management of temporally simultaneous EEG-
MREG recordings. Compared to non-simultaneous,
the simultaneous recordings have the advantage that
the two data types reflect the same neuronal
processes. This is because for both recordings the
condition of the subject is the same. Simultaneous
EEG-MREG recordings are used for instance in
localizing epileptic seizure. There are multiple
applications in the research area other than the
clinical one as researchers try to make a better
understanding of the neural processes. Makeig et al.,
(2002) and Czisch et al., (2004) are examples of
research papers that use simultaneous recordings for
clinical and developmental studies. Another paper is
the one by Jacobs et al., (2014). In their paper, they
analyse epileptic spikes from EEG-MREG to
determine the yield of fast MRI in the analysis of
intrinsic brain signals. A more recent paper is the
one by Rajna et al., (2015) detecting patterns of
brain activity by exploiting the superior spatial
accuracy of MREG data and the temporal dynamics
provided by EEG signals being 500 times faster than
MREG.
As the use of multimodal data such as EEG-
58
Fylakis, A., Keskinarkaus, A., Kiviniemi, V. and Seppänen, T.
Temporally Synchronized Reversible Data Hiding of EEG to MREG.
DOI: 10.5220/0005665700580067
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 5: HEALTHINF, pages 58-67
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

MREG with an increasing amount of details
recorded becomes more commonly used, efficiency
in data management becomes crucial. Efficiency
first refers to the storage and transfer of multimodal
data. The problem with multimodality is that when
data is transferred through the Internet and hospital
servers, there is risk of data loss due to the transfer
and storage of multiple files. Secondly, efficiency
refers to the high capacity of the data and the time to
access and analyse it. In our case, the use of EEG
and MREG as separate entities requires that access
to specific segments over time has to be done
separately. Also, as data is commonly stored
separately, it reserves more space and within
filesystems it requires that linkages between files
need to be handled manually.
Beyond the efficiency requirements are the
security requirements. Constant transfer of
biomedical data through networks and storage in
cloud databases raises security issues. Some among
the most important ones are confidentiality,
availability and reliability in biomedical data
(Coatrieux, et al., 2006). Concerning confidentiality,
biomedical data is private patient information and
thus direct access to all data by non-authorised users
would violate privacy. Availability refers to the
ability for direct access to all inconsistent entities.
Last, reliability has to do with the problems of
verifying integrity of data, as well as tracing and
validating authentic data. Tampered data can
mislead and cause errors in the diagnosis and similar
problems can occur when non authentic material is
used, so mechanisms for tracing and validating
authentic content are also required.
Data hiding has proved to significantly satisfy
those requirements by enriching data with metadata
and thus providing a new layer of security. Data
hiding is defined as the practice of imperceptibly
altering an object to embed a message about it (Cox
et al., 2007). In this case, we exploit data hiding
principles to firstly improve efficiency in data
management by embedding EEG signals in MREG
sequences. Secondly, confidentiality, availability
and reliability of data can be guaranteed. Access to
the hidden data can be restricted only to entitled
users, to ensure confidentiality. Then, as multimodal
data co-exist in a single package, availability is
ensured. Also, to improve reliability, hiding IDs or
digital signatures along host data can be used as a
proof of data’s ownership and authenticity (Grover,
1997) and in a similar manner, tamper-proofing is
solved. The target is to prove integrity with
embedded data and locate any possible tampering.
Embedded digital signatures in the host object are
capable after extraction and reversion of confirming
that no tampering has taken place and further use is
safe.
Integrity also requires reversibility in data hiding
techniques where modifications cause certain non
visible or slightly visible artefacts on host data. In
biomedical data, it is essential to use intact
information in the analyses. The solution comes with
reversible data hiding techniques. Reversible data
hiding refers to the recovery of the host medical
object to its exact initial state (Dharwadkar et al.,
2010).
Here, we propose a reversible data hiding
method based on histogram shifting. The original
histogram shifting method was proposed by Ni et al.
(2006) followed by improvements by Tsai et al.
(2009) and Fallahpour et al. (2011). The method
was implemented for 8-bit image applications; while
in this paper, we consider a multi-dimensional video,
that is, the 32-bit MREG as host. This is the main
novelty of this paper differentiating it from past
research which was focused on static images (Pan et
al., 2009). This paper further incorporates data
quantization in order to apply histogram shifting on
32-bit data.
Another novelty is temporal synchronization.
The EEG signal and/or other signals to be embedded
in the MREG sequences are synchronized in the
time domain. In practice, accessing an MREG
segment guarantees extraction of the simultaneously
recorded EEG. This is significant both time and
capacity wise as MREGs are big files requiring high
access time which is inefficient in the analyses.
Moreover, from the extracted EEG data, it is
possible to match samples from specific EEG
electrodes as they were embedded in specific order.
In medical imaging, the image can usually be
defined having two regions; the one of interest that
contains the imaged tissue and the variable extent
regions of no interest. A ROI (Region of Interest)
and a RONI (Region of Non Interest) can be selected
either by a medical doctor or automatically,
assuming for example that the background is of less
importance. There are unavoidably always voxels
outside the targeted tissue which have no meaning
for the diagnostic purposes. On the contrary,
distortions on the ROI area can cause errors in the
diagnosis. A novelty in the current method is the
option to restrict the histogram shifting idea to the
ROI, this way, elimination of the background in any
processing steps does not destroy hidden data, while
reversibility guarantees a recovery of the ROI.
The paper is organized as follows. Section 2
includes the description of the data hiding methods,
Temporally Synchronized Reversible Data Hiding of EEG to MREG
59
the data quantization, ROI segmentation and EEG
compression. Then, it includes the method to avoid
side information and the digital signature
exploitation. Section 3 explains the data formats and
the experimental results, Section 4 is the discussion
part of the paper and last Section 5 includes the
conclusion and suggestions for future work.
2 METHODS
The following subsections describe the embedding
and extracting-reversing methods of the proposed
technique. It can be described as the problem of
embedding a set of data w in a digital object I. Thus,
producing a new object I
w
, such that w can be reliably
located and extracted from I
w
(Collberg and Nagra,
2010), reversing at the same time I
w
to its original
state I. In this case, the message w is the EEG signal
and the host I is the MREG data where w is
embedded in I maintaining temporal synchronization
between I and w.
Not only reversibility is guaranteed, but also
imperceptibility of hidden data can be maintained
with specific settings related to data quantization
described in the following subsection. In our method
we hand data hiding using histogram shifting.
Because of the fact that the original method by Ni et
al. was designed for use in 8-bit images, which is
discrete data having a 256 bin histogram we had to
develop extensions that make use of data
quantization. This outputs a discrete histogram
applicable for the above scheme. MREG unlike
regular images is typically consisted of 32-bit data
and thus does not produce the common histogram of
an 8-bit image but a nearly flat histogram as values
are spread over 2
32
bins. For this reason the capacity
which is equal to the maximum histogram peak value
is limited.
Furthermore, another extension is a mode of
embedding only in the extracted important areas of
the image to increase robustness. This mode is also
tested and described in more detail within the
subsections that follow. EEG lossless compression
techniques are also suggested which could be of great
benefit in the extended mode where the ROI is
isolated because this restriction reduces capacity
significantly.
Following the algorithmic description of the
methods, subsection 2.5 presents a solution making
the extracting method blind, which means that it does
not require any side information as input. The last
subsection exploits the possibilities of digital
signature exploitation for integrity control, but also
as a measure to confirm the successful run of the
reversion along extraction.
2.1 Data Quantization
In all digital images, in order to form a digital
function, the gray-level values have to be converted,
i.e. quantized into discrete quantities. This process of
assigning gray levels to discrete intensity levels is
called quantization (Gonzalez and Woods, 2002).
The process of quantization can also refer in certain
cases to down-sampling existing discrete values.
Here, we deal with the second case as float single
precision values are actually discretised values but
with a very small quantization step.
In the MREG samples tested in this paper, image
data was as described in 32-bit single-precision
floating-point format, which means 2
32
histogram
bins while the original histogram shifting method
was optimized for 8-bit grayscale images and thus
for 2
8
histograms bins. A solution to solve this
format problem was to develop a method that down-
samples the histogram from 2
32
bins to 2
8
bins or
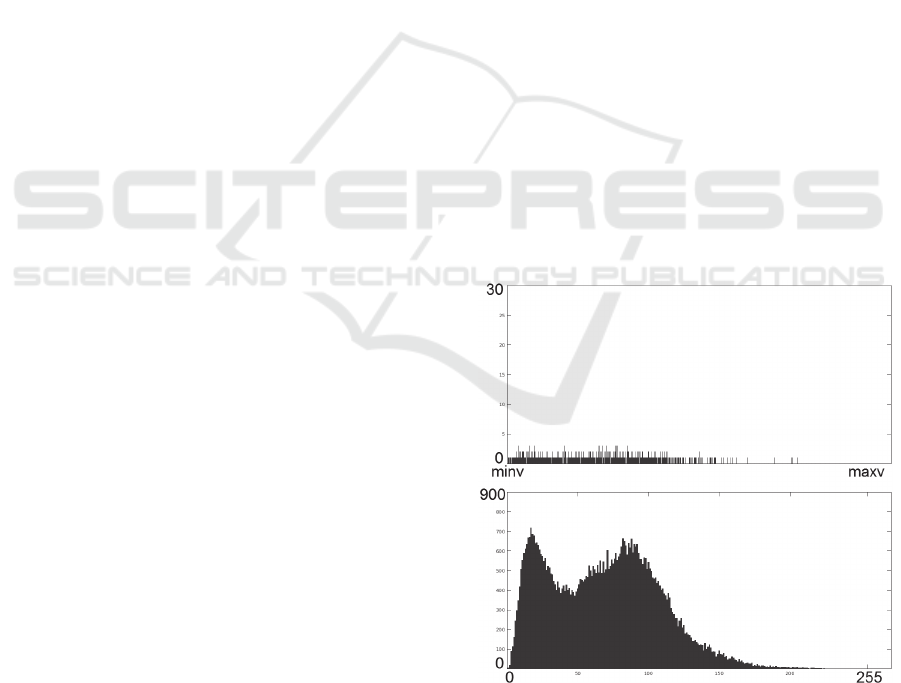
lower, as seen in Figure 1.
In our methods, we have tested varying data
quantization options getting histograms with less
than 256 bins in order to increase capacity,
something necessary for certain applications.
Specifically, results include tests from 256 down to
8 histogram bins.
Figure 1: Effect on histogram by exploiting quantization
getting down from 2
32
(top) to 256 bins (bottom).
Quantization should not downgrade image
quality, otherwise reversion is not possible. For that
purpose the quantization used in current methods
HEALTHINF 2016 - 9th International Conference on Health Informatics
60
can be better described as “grouping”. Thus, it does
not actually down-sample data but it creates
histogram bins, each one corresponding to an equal
range of data values. The quantization step is
calculated as follows:
Q = (maxv - minv) / bns, (1)
where maxv is the maximum intensity value, minv
the minimum intensity value and bns the target
number of histogram bins.
2.2 Restriction in the ROI
It is likely that modifications occur in the dark
background of the MREG, i.e. the RONI of the
MREG. For instance, voxel values of the
background can be removed or set to 0. Especially,
since this area outside the brain is usually extracted
from any analysis in order to increase the speed and
accuracy of brain analytics. Those modifications
would mean that data embedded there is lost without
making any visually distinctive difference to the
MREG. If we take the background of the brain as an
example, elimination of the whole background
would not make any visible or functional difference
for the analyses. In contrast, any modification in the
brain area affects the analyses, but those
modifications can be detected with methods using
digital signatures such as those described in
Subsection 2.6. As RONI elimination would affect
hidden data, we propose an extension restricting the
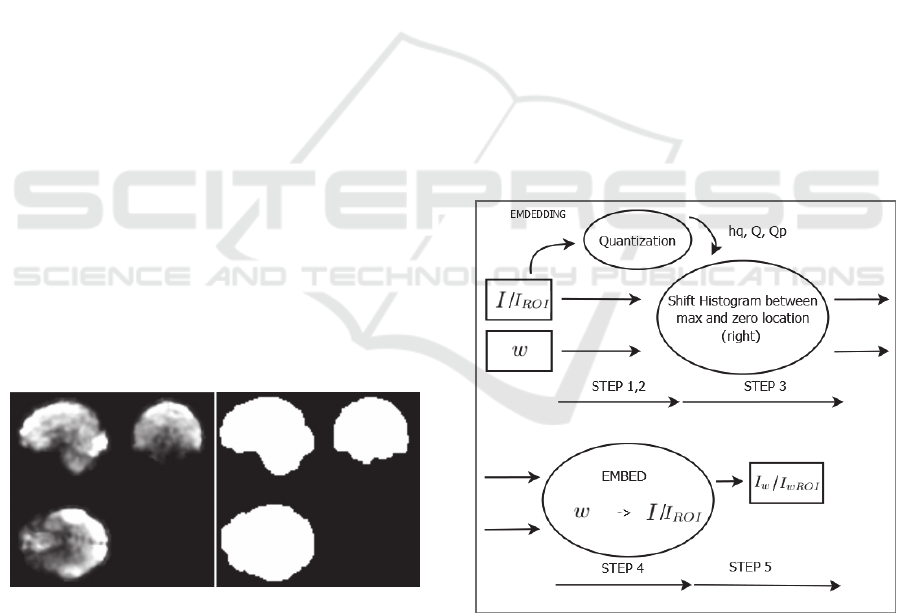
hiding target area in the ROI. For the ROI extracting
procedure, we use the BET2 brain estimation
algorithm by Jenkinson et al. (2005) creating a mask
of the imaged tissue, as Figure 2 depicts.
Figure 2: ROI masking using BET2.
2.3 EEG Compression
EEG lossless compression can also be incorporated
before embedding in the MREG to compensate
capacity loss caused by the segmentation and usage
as host of the ROI area only. Some recent examples
include the lossless multichannel compression
method proposed by Wongsawat et al. (2006) and
the lossless method making use of wavelet transform
and neural network predictors proposed by Sriraam
(2012). The above methods can achieve ratios of
2.77 to 1 and 2.99 to 1, respectively.
In the experiments presented in Section 3,
capacity comparisons can be made between
embedding raw and compressed EEG data using the
method proposed by Sriraam. All the experiments
have been performed embedding raw data, while
capacity using compressed EEG has been estimated
using the ratio referred at the paper.
2.4 Data Hiding and Extraction
The idea is applying the histogram shifting scheme
for an MREG sequence hiding an EEG or any other
physiological signal. That has to be done in a
temporal synchronized manner to make benefit of
the techniques that combine simultaneously sampled
recordings of EEG and MREG. Assume that the
sampling rates for EEG and MREG are s
1
and s
2
samples per second, respectively. In practice, that
means that for each MREG frame over time, the
embedding algorithm will hide data that corresponds
to F EEG samples.
F = s1 / s2 (2)
Figure 3: Embedding - block diagram.
The following embedding pseudocode describes
step by step a case with F EEG samples per MREG
frame where quantization is performed creating a
histogram with bns bins. An illustration is depicted
at the block diagram of Figure 3.
Temporally Synchronized Reversible Data Hiding of EEG to MREG
61
EMBED(w[L,B],I[M,N,P,T])
//Step 1
Q = GetQ(I,bns) (eq.1)
c←1
for vmin≤i≤vmax,i=i+Q
Qp[c]←i,c←c+1
//Step 2
for 1≤t≤T
hq[1…bns]←0
for 1≤i≤bns
for 1≤x≤M,1≤y≤N,1≤z≤P
if Qp[i]≤I[x,y,z,t]<Qp[i+1]
hq[i]←hq[i]+1
(mx,mxi)←max(hq) //(value,index)
(mn,mni)←min(hq) //(value,index)
//Step 3
for 1≤x≤M,1≤y≤N,1≤z≤P
if Qp[mxi+1]≤I[x,y,z,t]
if I[x,y,z,t]<Qp[mni]
I[x,y,z,t]←I[x,y,z,t]+Q
//Step 4
s←(t-1)*F+1 (eq.2)
for s≤l<s+F,1≤b≤B
(xi,yi,zi,t)←GetNextIdx(mxi)
if w[l,b]=1
I[xi,yi,zi,t]←I[xi,yi,zi,t]+Q
//Step 5
Iw←I
return Iw
END
First, the algorithm reads the input data w of L
samples over time, each of B bits, converted into a
binary stream and the MREG sequence I of size
MxNxP and temporal resolution T. I might either be
the whole MREG or the segmented ROI area
acquired as described in Subsection 2.2.
In Step 1, the quantization step Q is calculated,
given the MREG’s maximum and minimum
intensity values vmax and vmin and the number of
bins bns according to equation 1.
In Step 2, a loop for all the frames over time is
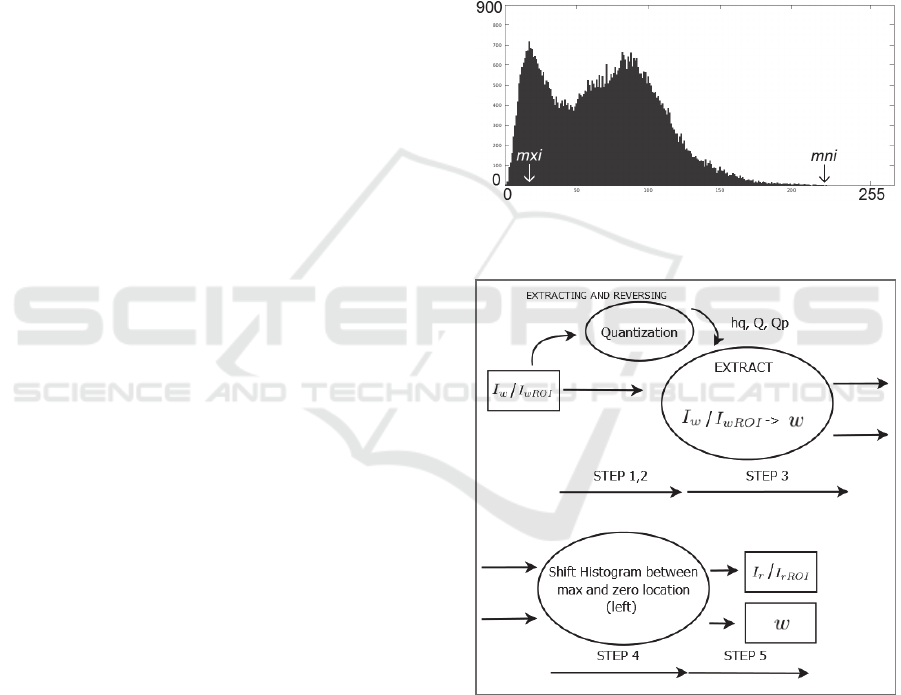
initiated and for each one the histogram Hq is
generated. Qp stores the positions of quantization’s
threshold points. Then, in the histogram Hq, the
maximum peak value and the minimum (zero) value
are located, which are hq(mxi), mxi ∈ [1,bns] and
hq(mni), mni ∈ [1,bns], respectively, as
demonstrated in Figure 4. Note that in case the
minimum value hq(mni) > 0, then, simply the
coordinates of those voxels and its greyscale values
are kept as overhead information to be hidden along
w and hq(mni) is set to 0.
In Step 3, assuming without loss of generality
that mxi < mni, bins following the peak up before the
zero location, i.e. hq(x), for every x ∈ (mxi,mni) are
shifted to the right by one histogram unit creating
one empty bin next to the maximum peak location’s
bin. This is performed by adding Q to each voxel
within this value range.
In Step 4, the algorithm accesses the voxels
which have intensity value within the range which
corresponds to the peak location histogram bin.
Voxel indexes in this range are returned sequentially
using function GetNextIdx(). As the binary
information to be embedded is accessed
sequentially, the voxel in order maintains its value to
store a 0 bit or is increased by the quantization step
value Q to store a 1 bit.
Finally, at Step 5, the modified MREG Iw is
returned.
Figure 4: Maximum peak location and minimum (zero)
location.
Figure 5: Extracting - block diagram.
For the extracting procedure, the MREG
containing hidden information Iw is entered as input
while the original maximum peak location and
minimum (zero) location are found without the need
of input side information. A solution for blind
extraction without side information is given in the
next subsection while the extracting method is
described in the following pseudocode and also
HEALTHINF 2016 - 9th International Conference on Health Informatics
62

illustrated at the block diagram of Figure 5.
EXTRACT(Iw[M,N,P,T])
//Step 1
Get Q,Qp (see, embed Step 1)
//Step 2
for 1≤t≤T
Get hq (see, embed Step 2)
Get mxi,mni (see, 2.5)
//Step 3
s←(t-1)*F+1 (eq.2)
for s≤l<s+F,1≤b≤B
(xi,yi,zi)←GetNextIdx(mxi)
if Qp[mxi]≤Iw[xi,yi,zi,t]
if Iw[xi,yi,zi,t]<Qp[mxi+1]
w[l,b]←0
if Qp[mxi+1]≤Iw[xi,yi,zi,t]
if Iw[xi,yi,zi,t]<Qp[mxi+2]
w[l,b]←1
Iw(xi,yi,zi,t)←Iw[xi,yi,zi,t]–Q
//Step 4
for 1≤x≤M,1≤y≤N,1≤z≤P
if Qp[mxi+2]≤Iw[x,y,z,t]
if Iw[x,y,z,t]<Qp[mni+1]
Iw[x,y,z,t]←Iw[x,y,z,t]-Q
//Step 5
Ir←Iw
return Ir, w
END
Following the same idea as in the embedding
method, data is extracted scanning the MREG
sequentially. Binary information is extracted
depending on whether greyscale voxel intensities
have retained the value corresponding to the
maximum peak location or they have been increased
by Q and thus moved to the next histogram bin.
MREG is reversed by returning those values back to
the peak location by subtracting intensities by Q and
shifting the histogram back by reducing the intensity
of the voxels corresponding to bins between the
maximum peak location mxi and the minimum
(zero) location mni by Q. Thus, the histogram is
shifted back to its original state and the MREG is
fully recovered. Last, the recovered MREG I
r
and
the extracted data w are returned.
2.5 Side Information
Side information refers to an extra input that the
extracting algorithm requires in order to perform its
operations. This input might be a secret key or some
information required for the extraction. In our case,
it refers to the maximum peak and minimum (zero)
histogram locations that have already been indexed
by running the embedding procedure.
In order to avoid the use of side information,
instead of detecting the location of the maximum
peak histogram value, the method detects the
location of maximum sum of consecutive pairs of
histogram values. Of course, this does not guarantee
detection of the right location. Thus, supplementary,
an identification code is also embedded in the first
bits along the payload, including the position of the
zero location. In case an invalid identification has
been extracted, then, the next largest pair of bins is
tested and so on until the right maximum peak and
minimum (zero) locations have been found.
Note that side information can be also used for
confidentiality purposes. Either the extracting
procedure is restricted being accessible only by
specific authorized users, or both the embedding and
extracting procedures require a secret key as side
information. Of course, this key is only known by
the authorized users (Cox et al., 2007). In this case,
the extracting procedure can be designed so that to
require the maximum peak and minimum (zero)
locations in the form of a key. Using an invalid key,
only irrelevant data is extracted.
2.6 Digital Signatures
For integrity control, an option is embedding a
digital signature of the original image along the
embedded payload. The digital signature will be
later proof as it is able to reveal data tampering.
Here, we use the Secure Hash Algorithm 2 (SHA-2)
(National Institute of Standards and Technology,
2014). It is a hash function designed by the U.S.
National Security Agency and published in 2001 by
the National Institute of Standards and Technology.
Different inputs produce different hashes and thus
different digital signatures for the input MREGs.
Here, the digital signatures are produced before
the embedding procedure, meaning that the original
MREG is used as input. After extraction and
reversion, the extracted signature is compared with
one produced from the reversed MREG. If the
signatures are identical, data integrity is confirmed.
This procedure is also useful in order to confirm that
reversion runs successfully. An invalid signature can
reveal errors of the process as the reversed MREG is
not identical to the original host object. This means
that it is not safe to use it for a diagnosis.
3 DATA AND EXPERIMENTS
For the experimental purposes, EEG data has been
collected through open databases available online at
physionet (Goldberger et al., 2000). All data samples
acquired had been already anonymized.
Temporally Synchronized Reversible Data Hiding of EEG to MREG
63

The MREG samples are also anonymous data
which was acquired from our university hospital
following research procedures with informed
consent.
In all experiments, the host MREG is consisted
by frames of 64x64x64 voxels, running time is 60
sec. and frame rate is 10 frames per second. Data is
in 32-bit single precision RAW NIfTI format.
The embedded EEG is 64 channel RAW data
with sampling rate of 160 samples per second. Each
data sample has 16-bit resolution. In total, in a one
minute run, we have 9,600 samples of 1,024 bits. Let
us note here that testing was not restricted to those
64 channels of embedded data but to the maximum
available capacity figures as well. This is performed
by simply repeating the available 64 channel data.
When ROI was segmented and used as input, the
capacity was significantly reduced as demonstrated
in the following subsection. In order to improve
those capacity figures, the quantization step Q is
increased accordingly, down-sampling data and
consequently decreasing the number of histogram
bins. This reduces marked images’ quality producing
even visible artefacts in certain cases, but
reversibility is guaranteed. Capacity is considerably
increased, but in some cases, it is clear that
compression of the EEG is also required to reach the
64 channel capacity of the testing set. That is why
lossless EEG compression techniques are applied.
The following subsections include information
about the capacity provided in the host MREG
thanks to the data hiding technique, as well as
information on data fidelity comparing the original
MREG with the one that carries hidden data.
3.1 Capacity
Table 1 shows the results acquired from eight
experiments over 1 minute of data measuring the
capacity both in bits of maximum available hiding
space, including standard deviation and in maximum
number of EEG channels that can be hidden.
Capacity refers to hosting capabilities per single
MREG frame. Note that temporal synchronization
was maintained meaning that for each MREG frame,
16 EEG samples over time are hidden.
At the first experiment, the whole MREG image was
used as a host. At the following six experiments, the
ROI of the MREG has been segmented restricting it
as the host area. In this case, different quantization
steps were tested, resulting from 256 to 8 histogram
bins. Restricting to the ROI also means that the
homogenous intensity of the image's background is
lost, reducing capacity significantly in a histogram
shifting technique. That is why higher quantization
is essential to get fair capacity figures. For example,
in our case where the EEG samples were consisted
of 64 channels, in order to get sufficient capacity
quantization step has to be significantly increased
while EEG compression is essential for smaller
quantization steps.
Table 1: MREG’s capacity.
Host area,
# bins
Capacity
# bits
Mode1
# channels
Mode2
# channels
Entire, 256 187958±0.49 734 2185
ROI, 256 888±0.00 3 10
ROI, 128 1366±26.29 5 15 – 16
ROI, 64 2672±42.91 10 30 – 31
ROI, 32 5264±63.09 20 60 – 61
ROI, 16 9846±99.79 38 113 – 115
ROI, 8 18126±185.03 70 – 71 208 – 212
The impact of compressing EEG data is depicted
as follows: Mode 1 shows the number of embedded
uncompressed EEG channels, while Mode 2 the
number of compressed channels estimated according
to the Sriraam’s (2012) method in which a
compression ratio of 2.99 can be achieved.
3.2 Fidelity
For the perceptual quality experiments, Table 2
includes the fidelity figures for the same tests
performed in the previous subsections. In every case,
a 1 minute EEG signal segment was hidden in a 1
minute MREG segment maintaining temporal
synchronization. In the first row, once again, the
entire image was used as host, while at the second
row and below, the ROI was segmented and used as
host. Gradually, the quantization step is increased
and thus the number of histogram bins is decreased.
In every case, the figures include the Peak Signal
to noise ratio (PSNR) for exploitation of the
maximum number of channels that capacity enables,
as seen on Table 1. Input MREGs for the PSNR
function were down-sampled to 8 bits. Concerning
the current test set and being restricted to 64 EEG
channels, PSNR was 51.25 dB using the entire
MREG as host and 256 bins and 30.42 dB using the
segmented ROI as host and 8 histogram bins.
Figure 6 demonstrates the result of embedding 64
EEG channels in 256 bins at the entire image, while
Figure 7 shows another case of embedding 10 EEG
channels using the ROI and the maximum available
capacity when 64 bins are produced. Last, Figure 8
depicts an extreme case of embedding 64 EEG
channels in 8 bins of the ROI.
HEALTHINF 2016 - 9th International Conference on Health Informatics
64
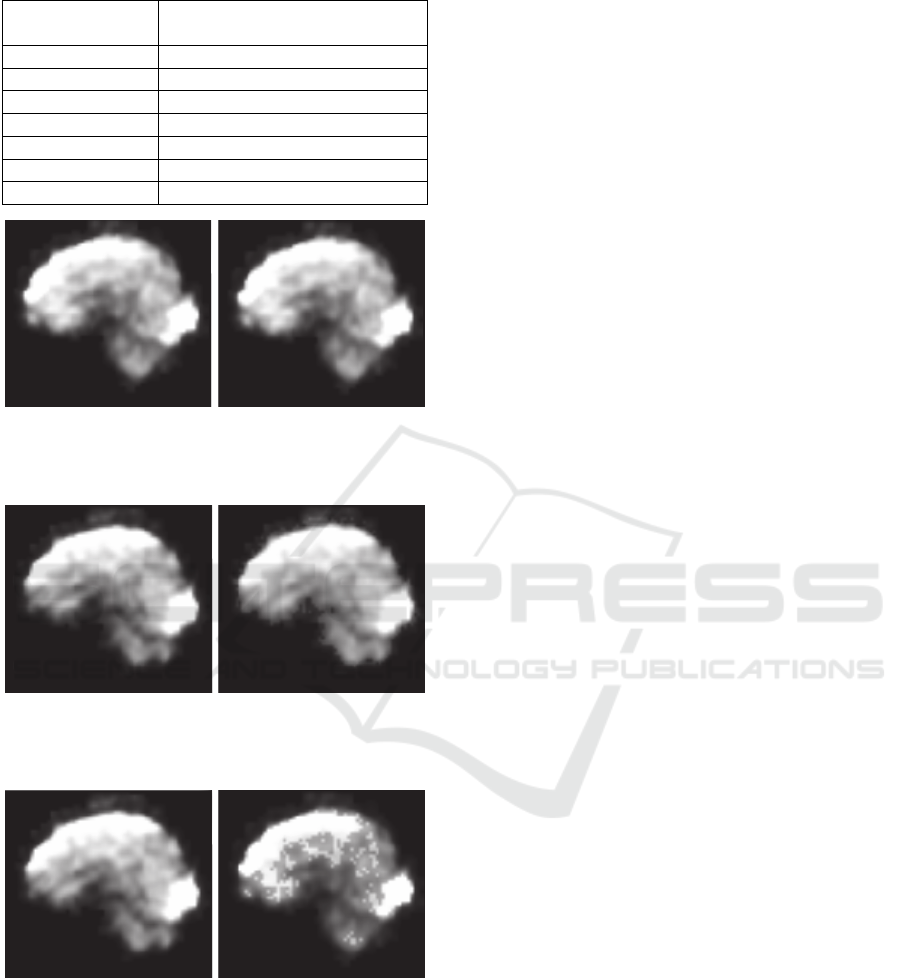
Table 2: MREG’s fidelity.
Host area, # bins
PSNR (dB) for max. # channels
Entire, 256 53.34
ROI, 256 78.34
ROI, 128 48.94
ROI, 64 42.79
ROI, 32 36.82
ROI, 16 32.92
ROI, 8 30.54
Figure 6: Original (left) and marked (right) MREG
intersection (64 EEG channels, Entire MREG, 256 bins,
PSNR: 51.25 dB).
Figure 7: Original (left) and marked (right) MREG
intersection (10 EEG channels, MREG’s ROI, 64 bins,
PSNR: 42.79 dB).
Figure 8: Original (left) and marked (right) MREG
intersection (64 EEG channels, MREG’s ROI, 8 bins,
PSNR: 30.42 dB).
4 DISCUSSION
Nowadays, it is very common for medical data to be
stored in clouds and to be transferred between
databases through open hospital networks. A typical
example of such an application can be seen in
biobanking. Biobanks are electronic repositories
where big data collections with relevant consents of
the donors are stored and furthermore made
available to research through common availability
services.
In biobanking applications, efficiency in using
the vast amounts of increasing data, as well as the
fact that there might always be disputes over
originality and authenticity of data creates big issues
(Ruzzo, 2014). Furthermore, it is expected that a
great amount of development in applications and
research related to information collected in biobanks
will take place in the near future. All things
concerned provide a solid motivation towards the
research in data hiding with applications in
biomedical databases.
The main novelty of the paper is the introduction
of a reversible data hiding method for a
multidimensional data sequence that is the MREG.
To our knowledge, this is the first paper to feature
the temporal synchronization of EEG and MREG
recordings. Past literature focuses on static
biomedical images; a thorough review of which is
presented by Pan et al. (2009).
In the introduction, the paper points out
numerous benefits of utilizing the data hiding
method in EEG-MREG applications. It is
demonstrated that, in general, packaging MREG and
EEG in one entity is very significant for efficiency
in data management and storage. Moreover, it
should be pointed out that this is also beneficial for
the visualization of MREG sequences combined
with EEG data. The existence of temporally
synchronized data in one package enables the
potential for significant visualization improvements
as there can be instant view of the corresponding
EEG for a given MREG sequence segment over
time. A user can select an MREG segment to view
or analyse, and then by the use of the data hiding
method, the user may also efficiently acquire an
output of the reversed MREG data accompanied
with the temporally equivalent EEG signal’s
segment. So far, this is a process that has been done
manually, and thus our method demonstrates data
hiding’s benefits on data management efficiency in
visualisations.
In every experiment, the embedded data was 16-
bit EEG data of 64 channels and 160 samples per
second but properties can vary. For different cases,
capacity figures can be easily approximated thanks
to the column that shows the capacity in number of
bits in Table 1.
The proposed data hiding method is reversible,
Temporally Synchronized Reversible Data Hiding of EEG to MREG
65

so the original data can always be retrieved.
However, with the tuneable quantization stepsize
parameter, the fidelity of the MREG sequences
carrying the EEG signal can be controlled. Best
quality figures are attained with low down-sampling.
ROI segmentation reduces capacity so different
quantization stepsizes were tested finding out that
fair fidelity is maintained with the use of 64
histogram bins in embedding or higher. In those
cases, PSNR was maintained over 40 dB which is
generally considered a threshold when it comes to
imperceptibility.
There are certain cases where the image requires
absolute fidelity which practically means that it
should be visually identical to the original MREG
throughout all its phases of use. This also concerns
usage before data is extracted and reversed for the
analyses’ purposes. For instance in applications
requiring preview of the MREG, it is important that
higher quantization is avoided, and thus ROI
segmentation should preferably not be considered.
Otherwise visible artefacts which can be disturbing
might appear on the MREG. Alternatively, EEG
compression can be a solution for this problem
because lower quantization can be enough for the
required capacity.
5 CONCLUSIONS
This paper presented a method for hiding EEG or
other physiological signals into MREG with a main
purpose of providing efficiency in data management
and storage. Furthermore, the paper addressed
security issues, i.e. confidentiality, availability and
reliability of content. Tamper proofing capabilities
are additionally provided as small alterations on the
host image affect hidden data and thus illegitimate
extracted data or digital signatures imply data
tampering. The data hiding method can guarantee
with the proper quantization settings high fidelity
between the original MREG sequence and its
version that carries hidden data. Moreover
reversibility is available. Along extraction of data,
the method reverses the MREG that carries hidden
data to its original state. Last, temporal
synchronization between EEG and MREG data is
always maintained.
In future work, we will develop methods for
increasing data hiding capacity. The purpose is to
combine more data modalities hosted in medical
data.
REFERENCES
Collberg, C., and Nagra, J., 2010. Surreptitious software.
Addison-Wesley, Upper Saddle River, NJ, 1
st
edition.
Coatrieux, G., Lecornu, L., Sankur, B., and Roux, C.,
2006. A review of image watermarking applications in
healthcare, In Engineering in Medicine and Biology
Society, 28th Annual International Conference of the
IEEE, pages 4691–4694.
Cox, I., Miller, M., Bloom, J., Fridrich, J., and Kalker, T.,
2007. Digital watermarking and steganography.
Morgan Kaufmann. Burlington, MA, 2
nd
edition.
Czisch, M., Wehrle, R., Kaufmann, C., Wetter, T. C.,
Holsboer, F., Pollmacher, T., and Auer, D. P., 2004.
Functional MRI during sleep: BOLD signal decreases
and their electrophysiological correlates. In European
Journal of Neuroscience, volume 20, pages 566–574.
Dharwadkar, N. V., Amberker, B., Panchannavar, S., and
Panchannavar, P., 2010. Reversible fragile medical
image watermarking with zero distortion. In Int’l
Conference in Computer and Communication
Technology, IEEE, pages 248–254.
Fallahpour, M., Megias, D., and Ghanbari, M., 2011.
Reversible and highcapacity data hiding in medical
images. In IET Image Processing, 5(2):190–197.
Goldberger, A. L., Amaral, L. A., Glass, L., Hausdorff, J.
M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody
G. B., Peng C. K., and Stanley, H. E., 2000.
Physiobank, physiotoolkit, and physionet components
of a new research resource for complex physiologic
signals. In Circulation, 101(23):215–220.
Gonzalez, R. C., Woods, R. E., 2002. Digital image
processing, Prentice-Hall. Upper Saddle River, NJ, 3
rd
edition.
Grover, D., 1997. The protection of computer software -
its technology and applications, Cambridge University
Press. NY.
Jacobs, J., Stich, J., Zahneisen, B., Assländer, J.,
Ramantani, G., Schulze-Bonhage A., Korinthenberg,
R., Hennig, J., and LeVan, P., 2014. Fast fMRI
provides high statistical power in the analysis of
epileptic networks. in Neuroimage, volume 88, pages
282–294.
Jenkinson, M., Pechaud, M., and Smith. S., 2005. BET2:
MR-based estimation of brain, skull and scalp
surfaces. In Eleventh Annual Meeting of the
Organization for Human Brain Mapping, volume 17.
Makeig, S., Westerfield, M., Jung, T. P., Enghoff, S.,
Townsend, J., Courchesne, E., and Sejnowski, T. J.,
2002. Dynamic brain sources of visual evoked
responses. In Science, volume 295, pages 690–694.
Menon, V., and Crottaz-Herbette, S., 2005. Combined
EEG and fMRI studies of human brain function. In
International Review of Neurobiology, volume 66,
pages 291–321.
National Institute of Standards and Technology, 2014.
Descriptions of SHA-256, SHA-384, and SHA-512.
csrc.nist.gov/groups/STM/cavp/documents/shs/sha256
-384-512.pdf.
HEALTHINF 2016 - 9th International Conference on Health Informatics
66

Ni, Z., Shi, Y. Q., Ansari, N. and Shu, W., 2006.
Reversible data hiding. In IEEE Transactions on
Circuits and Systems for Video Technology,
16(3):354-362.
Pan, W., Coatrieux, G., Montagner, J., Cuppens, N.,
Cuppens, F., and Roux, C., 2009. Comparison of some
reversible watermarking methods in application to
medical images, In 31st Annual International IEEE
Conference on Engineering in Medicine and Biology
Society, pages 2172–2175.
Rajna, Z., Kananen, J., Keskinarkaus, A., Seppänen, T.,
and Kiviniemi, V., 2015. Detection of short-term
activity avalanches in human brain default mode
network with ultrafast MR encephalography, In
Frontiers in Human Neuroscience, 9(448).
Ruzzo, A., 2014. Biobanking's big problem: how do you
manage all that data? blog.fisherbioservices.com/bid
/376102/Biobanking-s-Big-Problem-How-do-you-
manage-all-that-data.
Sriraam, N., 2012. A high-performance lossless
compression scheme for EEG signals using wavelet
transform and neural network predictors. In
International Journal of Telemedicine and
Applications, 2012(5): 1–8.
Tsai, P., Hu, Y. C., and Yeh, H. L., 2009. Reversible
image hiding scheme using predictive coding and
histogram shifting. In Signal Processing, 89(6):1129–
1143.
Wongsawat, Y., Oraintara, S., and Rao, K. R., 2006.
Integer sub-optimal Karhunen-Loeve transform for
multichannel lossless EEG compression. In 14
th
European Signal Processing Conference, IEEE, pages
1–6.
Temporally Synchronized Reversible Data Hiding of EEG to MREG
67
