
ZK DrugResist
Automatic Extraction of Drug Resistance Mutations and Expression Level
Changes from Medline Abstracts
Zoya Khalid
1
and Osman Ugur Sezerman
2
1
Department of Biological Sciences and Bioengineering, Sabanci University, Istanbul, Turkey
2
Department of Biostatistics and Medical Informatics, Acıbadem University, Istanbul, Turkey
Keywords: Drug Resistance, Mutations, Gene Expression, Naive Bayes, Machine Learning.
Abstract: Drugs are small molecules that generally work by binding to its target which is often a protein. This ligand
molecule binding helps in the treatment of various diseases. Major obstacle to treat complex diseases is the
phenomena underlying drug resistance mechanisms which are not fully understood so far. Previously reported
literature has mentioned few of the motives behind this complex mechanism which dominantly include protein
missense mutations and the changes in the expression levels of certain genes. A better understanding of these
mechanisms is getting crucial for the researchers. Retrieving information on these processes can be
challenging as scientific literature has huge pool of data and extracting the required information has always
been a laborious task. We developed an online pipeline ZK DrugResist that automatically extracts PubMed
abstracts of drug resistance paired with either mutation or expression for a given disease. Our classifier
showed 97.7% accuracy with 93.5% recall and 96.5% F-measure. This system saves plenty of time in terms
of data mining and also reduces efforts in retrieving information from online resources.
1 INTRODUCTION
The term drug also referred as dose or medication is
used for treatment of various diseases. There are two
ways to classify drugs, one named as the small
molecule drugs which include proteins, biological
medicinal product and vaccines which further used as
therapeutic agents for the treatment of certain
diseases. The second way of classification is based on
how the drug is administered that is its specific mode
of action following the therapeutic effects. The drug
usually functions by binding to its target which is
often a protein. Proteins are large biomolecules made
up of amino acids. They are also visualized as large
globular structures that have deep groves in it, which
may have buried binding site that is good for
druggability. The drug molecule will then fits in the
binding site and the process is termed as ligand-
molecule binding. In this way the drug performs its
action and helps diagnosing and curing various
diseases (Dean et al., 2005; Michael, 2002; Walsh,
2000).
Sometimes treatment phase has been passed
through an obstacle “drug resistance” generally
meaning the decrease in the efficacy of the drug in
curing a disease. This is the major constraint to treat
complex diseases. The underlying mechanisms are
not very clear but still there are some notions about it.
First theory states that drugs at their certain target
sites are present in a decreased concentration caused
by increased level of expression of drug molecules.
Second involves the modification of drug targets
which affects the protein-ligand binding complex
(Remy et al., 2003). Drug resistance has a strong
impact on disease treatment; it has been observed that
in many of the cases this brings failure in treatment.
This shows that rate of survival is proportional to how
strongly the mechanism of drug resistance is being
overpowered. The survival chances would increase if
the drug resistance could be overcome (Longley and
Johnston, 2005).
The current study focuses on evaluation of
complex phenomena lying behind the drug resistance
mechanism. From the literature it has been found that
one of the major reasons behind this is the protein
alteration which involves amino acid mutations at
certain residue. Theses missense mutations affect the
binding affinity of the protein with the ligand and
hence results in making drug insensitive to the
168
Khalid, Z. and Sezerman, U.
ZK Drugresist - Automatic Extraction of Drug Resistance Mutations and Expression Level Changes from Medline Abstracts.
DOI: 10.5220/0005664501680173
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 3: BIOINFORMATICS, pages 168-173
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

treatment. For example as reported in previous
studies that V299L, T315A, and F317I/L mutations
are resistant against dasatinib while mutations
likeY253F/H, E255K/V, and F359C/V are resistant
for nilotinib, therefore making protein mutations as
an important factor for drug resistance mechanisms
(Chrisanthar et al., 2008; Hochhaus et al., 2011).
Second important factor is the expression based drug
resistance mechanism. The changes in expression
level which either is the overexpression or down-
regulation of certain genes induces enhanced
resistance against various drugs. As one of the studies
reported the overexpression of ANP32C creates
enhanced resistance against FTY720 drug, hence
makes it ineffective to treatment (Buddaseth et al.,
2014).
In order to retrieve and comprehend drug
resistance mechanisms, researchers either has to look
for the online databases or read all freely available
biomedical documents through online sources, which
is of course a very time consuming task. Many
computational biology/bioinformatics studies have
focused in building automated pipelines to extract
information from PubMed abstracts. There are some
databases published in literature that stores different
aspects of drug and gene relationship like BacMet
which focuses on genetic alterations causing
resistance against antibiotics (Pal and Larsoon, 2014).
Moreover there is another tool named Biozyne P-gp
Predictor which is based on SVM classifier that
differentiates the substrates from efflux pumps
(Levatic et al., 2013). Another similar reported
database is CancerDR which focuses on the
identification of the altered genes encoding drug
targets (Kumar et al., 2013). Retrieving information
from such kind of repositories is a laborious task.
Making automated way of information retrieval is one
solution to this. Previously published methods just
focussed on general analytical tasks like mining genes
and protein names or describing relationship of genes
and drug. These methods don’t emphasize on
combining all these information and placing them in
one platform. Some of the tools on information
mining are already been published, for instance Proux
research group reported the syntactic parsing
methodology for information extraction developed by
(Proux et al., 1998). Similarly another method used
statistical based information (Hishiki et al., 1998;
Ohta et al., 1997). In the same way (Cutting and
Kupiec, 1992; Aronson et al., 1994; Humphreys et al.,
1998 ;) also developed servers that used semantic
analysis approach for information extraction.
Thorough review of the literature revealed that there
is another published tool EDGAR (Rindflesch et al.,
2009) that overcomes the limitations of the previously
existing information retrieval methods. This tool
works in building relationship between genes and
drugs relevant to cancer therapy. But unfortunately
this tool is not available online yet and it is also not
mentioned that how much accuracy authors have
achieved in applying natural language processing on
the abstracts. In another study reported by (Bui et al.,
2010) the authors developed the method for
combining drug and mutation level information for
HIV. Again this method is only specific to HIV. Our
proposed method has successfully benchmarked
already existing methods. ZK DrugResist uses
machine learning approach to retrieve drug resistance
information. It provides one platform that gathers
gene names, drug names, abstracts titles, link to the
abstracts categorised by disease type. Our tool
provides the most systematic way of information
extraction for drug resistance abstracts available on
PubMed. In this way it facilitates the researchers in
mining desired information more robust and more
accurate.
The PubMed directory considered as a rich source
of information as it has a huge collection of abstracts.
Despite this fact, automated mining of worthy
information remains a big challenge for researchers.
Our study aims to develop an online tool to
automatically extract all the abstracts from PubMed
related to drug resistance. These abstracts and the
related information are downloaded into a database.
From this all the information about the mutation, gene
and the expression status is processed and displayed
on web. Furthermore the abstracts are also marked as
cancer or other diseases based on the content
provided in the abstract. We used MugeX and
EnzyMiner approach developed by our
computational biology group for implementing this
classifier (Erdogmus and Sezerman, 2007; Yeniterzi
and Sezerman, 2009).
2 METHODOLOGY
Abstracts available online queried by using search
terms: “Drug resistance”, “amino acid mutation at
drug resistance level”, “expression based drug
resistance” and different combinations of these terms
were downloaded from Medline which are many
thousands in number. Out of them only those
abstracts are filtered that has either the drug resistance
and the protein mutations content present together or
drug resistance and the expression level information
present in a document. The downloaded abstracts
were passed through variety of algorithms including
ZK Drugresist - Automatic Extraction of Drug Resistance Mutations and Expression Level Changes from Medline Abstracts
169

tokenization/sentence splitting followed by porter
stemming. These algorithms are applied in order to
break down the abstract into sentences and then into
words making them easy to process.
2.1 Classification Modules
We applied two learning algorithms Naive Bayes and
Rocchio algorithm both uses bag of words approach.
After pre-processing the dataset, we applied our
classifiers which for our case are based on four levels
of classification. First stage is to separate the abstracts
of drug resistance from the other ones. First the
document is processed by tokenization and porter
stemming algorithm. Further we applied TF-IDF
weighting (term frequency inverse document
frequency). It is the product of two statistics Term
Frequency and Inverse Document Frequency, term
frequency deals with the raw calculation of a term in
a document while inverse document frequency deals
with the significance of a word count. We observed
how many times word “Drug Resistance” appears
together in a document, if it is more common we
labelled the document as drug resistance else it is
labelled as others. Following this the next phase
classification picks the drug resistance tagged
abstracts to further classify them as either mutation or
expression. The regular expressions are designed for
this purpose. If regular expression matches any
mutation related information in the document we
marked it as mutation, on the other hand it is marked
as expression based if the content displays the gene
expression level changes for the drug resistance again
TF-IDF is used for this purpose. Third step is to sub-
categorize the abstracts labelled as mutations. The
mutation can be at amino acid level or at the
nucleotide level our tool is interested only to pick the
protein mutations. Those at DNA level are termed as
ambiguous mutations, so this step targets to remove
ambiguous mutations from the actual ones using the
regular expressions defined earlier. The last module
of our classifier is to divide the cancer related articles
with the ones which are showing other diseases of
metabolic, autoimmune and neurodegenerative. The
documents cited by terms cancer, leukaemia and
tumour belongs to cancer class while the rest are
classified in others category. For term frequency we
used TF-IDF as mentioned before. Figure 1
summarizes all the steps involved in ZK DrugResist.
For implementation Perl Regular Expressions
were used, set of patterns were formed describing the
protein mutations, for instance the mutation cited as
L15V, Arg567Leu, Ala399->Asp and some are
mentioned as full sentences substitution of
Methionine with Valine at position 40. Following the
mutations the gene names parallel to mutation stated
in the abstracts were also downloaded and stored in
the database.
2.1.1 Drug Resistance Vs. Others
As mentioned first stage of classification is to clearly
mark the abstracts which are showing drug resistance
mechanism from the others which are irrelevant to
these. The total abstracts downloaded are 701 in
number. For each of the downloaded abstract the
feature vector is constructed. In order to distinguish
them the frequency of each word is counted as a
feature value. These words were then further
processed using tokenization and porter stemming
algorithms. After breaking the abstract into words and
counting the frequency of term “Drug Resistance” it
is marked as either drug or others.
2.1.2 Mutation Vs. Expressions
In the second category we picked these drug
resistance documents and scanned them for the
mutation level information. The documents cited
using overexpression down regulation kind of terms
are marked as expression abstracts while those which
uses amino acid terminology are marked as mutation
by our algorithm.
2.1.3 Protein Vs. DNA
The Perl regular expressions were applied to extract
the amino acid level mutations from each document.
The major hindrance is the mixing of some protein
mutations with the DNA ones. For example the one
letter code amino acid mutation like A456G can
easily be misinterpreted with the nucleotide letters.
We compiled regular expressions for this ambiguity
to be solved. The documents are classified based on
the content information.
2.1.4 Cancer Vs. Others
In the last module of classification those abstracts
which are associated with cancer were separated with
the abstracts which are related to other diseases which
include neurodegenerative, autoimmune and
metabolic disorders.
2.1.5 Gene/Inhibitors
The gene names following the protein mutation are
also extracted from the abstracts. For this purpose the
complete list of official gene names were being
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
170

downloaded from HUGO database
http://www.genenames.org/. Any gene name
mentioned in the abstract is programmed to match
with the list of the genes stored and the results are
displayed on web. We followed MugeX approach for
this module.
2.2 Implementation
All the steps are implemented in The Perl
Programming Language. Strawberry Perl version
5.20 was used. The regular expressions were
compiled using PERL Regular Expression library.
The necessary information from the articles including
Title, Abstract and PubMed ID was downloaded and
stored in XML format into MySQL database. After
this the documents are processed first using
tokenization using Perl module PPI::Tokenizer. Each
sentence is broken into words and further porter
stemming was applied to each document. This
algorithm is used to remove the common words from
English which are actually not contributing in
classification for example “the”, “is”, “are” and
similar words to that. After pre-processing all the
documents, they are passed through Naive Bayes and
Rocchio classifiers in order to achieve four levels
classification. The database is built on Xampp Server;
database tables are stored in MySQL phpmyadmin of
xampp. The web interface was designed in
WordPress using html and PhP. CGI, DBI and DBD
modules of Perl were being used for retrieving the
data from MySQL databases and displaying the
output on the web page.
2.3 Testing
In order to test the classification results training,
testing and k fold cross validation were employed.
For all the four modules of classification 20% of the
abstracts were being used as test set, while remaining
abstracts were considered as training set. We
performed 5 fold cross validation that means the
whole data is being divided into 5 sets out of which 4
are used are training sets and the rest of one is as test
set. These sets are being shuffled 50 times and
average accuracy for both the training and test sets
were being measured.
3 RESULTS
The classifier is tested with the entire pre-processing
algorithms we implemented. Out of huge pool of data
available online on drug resistance mechanisms we
only filtered those which are showing either mutation
level or expression level changes in causing drug
resistance. This makes up to 701 documents in total.
The first module of classification separates out 144
documents as drugs while the other 557 are the ones
in which mutation is mentioned but not at drug level.
These 144 documents are advanced to the second
level of classification. This shows that out of 144
documents, 91 are those belonging to mutation
category, 22 of them are the expression based
resistance abstracts while rest of 31 did not show
either the mutation or the expression based drug
resistance. These 91 abstracts are then picked to
distinguish the protein and DNA level mutations from
each other. The results showed that 65 of them are the
ones which are being labelled as amino acid
mutations while 25 are the other ones. Last module of
classification separates the cancer disease ones with
the other diseases. The calculation shows that 53 are
the drug resistance mutations at cancer level while 12
are the ones which are in other category. We
compared the results of our classifiers and the results
showed that Naïve Bayes classification outperformed
Rocchio algorithm in precision, recall and accuracy
as shown in Table1. The results of Naive Bayes
classifier are listed in Table 2, 3, 4 and 5 respectively.
The graphical representation is illustrated in Figure 1.
ZK DrugResist is a user friendly web application,
every time a user queries to find the mutations or the
expression based drug resistance information, the in-
built program connects to the MySQL database tables
and displayed the output on webpage as shown by one
of the snapshot in Figure 2. The classifier shows 97%
average accuracy on test set for 5 fold cross
validation.
Table 1: Comparison of Naive Bayes and Rocchio
Algorithm.
Naive Bayes Classifier Rocchio Algorithm
Accuracy Recall Precision Accuracy Recall Precision
97% 96.5% 95.9% 90.5% 83.0% 89.4%
Table 2: Classification Results of Drug Resistance vs.
Others in Training and Test Sets.
No of
Abstracts
Accuracy
Measure
Recall F-measure
660 96.4% 95.4% 93.7%
140 96.7% 96.5% 95.9%
ZK Drugresist - Automatic Extraction of Drug Resistance Mutations and Expression Level Changes from Medline Abstracts
171
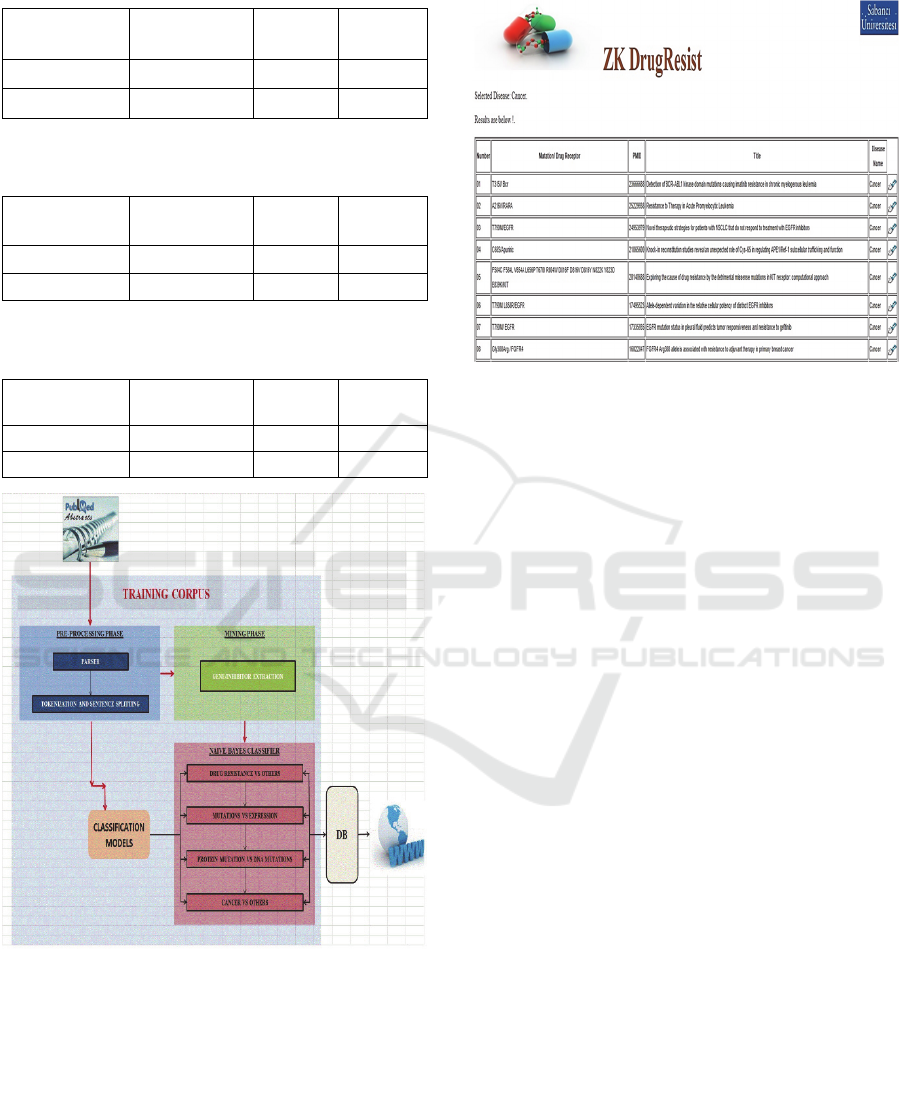
Table 3: Classification Results of Mutations vs.
Expressions in Training and Test Sets.
No of
Abstracts
Accuracy
Measure
Recall Precision
115 96% 95.4% 93%
28 97% 96.5% 95.9%
Table 4: Classification Results of Protein Mutations vs.
DNA Mutations in Training and Test Sets.
No of
Abstracts
Accuracy
Measure
Recall Precision
72 96% 94% 93%
18 96.7% 96.5% 95.9%
Table 5: Classification Results of Cancer vs. Others in
Training and Test Sets.
No of
Abstracts
Accuracy
Measure
Recall Precision
52 96% 94% 93%
13 96.7% 96.5% 95.9%
Figure 1: Flowchart of Methodology.
4 CONCLUSIONS
In this study we developed an online pipeline ZK
DrugResist to find the PubMed abstracts of drug
resistance combined with protein and expression
level data. Our tool outperformed the already existing
tools. ZK DrugResist is very proficient in mining
drug resistance semantics in an automated way from
Figure 2: Snapshot of ZK DrugResist showing the abstracts
from cancer disease.
literature without comprising the accuracy measure.
It is freely available online and is a self-explanatory
tool aimed to help researchers in finding desired
information on one click. As for the future work we
might extend this tool to also work on full articles
rather just on the abstracts. As in some cases we found
that the desired information is missing in the abstract
but present in the remaining article. This will further
increase our dataset size and may also improve the
accuracy measure. Our tool is available
at
http://zkdrugresist.sabanciuniv.edu/.
REFERENCES
Aronson, A, Rindflesch, T & Browne, A 1994, ‘Exploiting
a large Thesaurus for information retrieval’, in
Proceedings of RIAO, pp. 197-216.
Buddaseth, S, Gottmann,W, Blasczyk & Huyton, T 2014,
‘Overexpression of the pp32r1 (ANP32C) oncogene or
its functional mutant pp32r1Y140H confers enhanced
resistance to FTY720 (Finguimod)’ , Cancer Biol Ther,
vol.15, no. 3 ,pp. 289-96.
Bui, Q, Nuallan, B, Boucher, C & Sloot, P 2010,
‘Extracting Casual Relations on HIV drug resistance
from literature’, BMC Bioinformatics, vol.11, no.101.
Chrisanthar, R, Knappskog S, Lokkevik, E, Anker G
,Ostenstad, B, Lundgren,S, Berge, EO, Risberg, T,
Mjaaland, I, Maehle, L, Engebreston, LF, Richard, J,
Lillehaug &Loning, PE 2008, ‘CHEK2 Mutations
Affecting Kinase Activity Together With Mutations
in TP53Indicate a Functional Pathway Associated with
Resistance to Epirubicin in Primary Breast Cancer’,
PLoS ONE, vol. 3, no. 8, pp. e3062.
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
172

Cutting, D & Kupiec, J 1992, ‘A practical part of speech-
tagger’, In proceedings of Third conference on Applied
Natural Language Processing, pp.133-140.
Dean, M, Fojo, T & Bates, S 2005, ‘Tumour stem cells and
drug resistance’, Nature Reviews cancer, vol.5, no. 4
pp.275-284.
Erdogmus, M & Sezerman, U 2007, ‘Application of
Automatic Mutation-gene Pair Extraction to Diseases’,
J Bioinform Comput Biol, vol. 5, no .6, pp.1261-1275.
Gottesman, M 2002, ‘Mechanisms of cancer drug
resistance’, Annual review of medicine, vol. 53, pp.615-
627.
Hishiki,T, Collier, N, Nobata, C, Okazaki, T, Ogata,
N,Sekimizu, T, Steiner, R, Park, H & Tsujii, T 1998,
‘Developing NLP tools for Genome Informatics: An
Information Extraction Perspective’, Genome Inform
Ser Workshop Genome Inform, pp. 81-90.
Hochhaus, A, Rosee, PL, Muller, MC. Ernst, T & Nicholas,
CP 2011, ‘Impact of BCR-ABL mutations on patients
with chronic myeloid leukemia’, Cell cycle, vol. 10, no
.8, pp. 250-60.
Humphreyes, B, Lindberg, D, Schoolman, H & Barnette, G
1998, ‘The Unified Medical Language System; an
informatics research collaboration’, J Am Med Inform
Assoc, vol. 5, no. 11, pp.1-11.
Kumar, R, Chaudhay, K, Gupta, S, Singh, H, Kumar, S,
Gautam, A, Kapoor, P & Raghhava, GP 2013, ‘Cancer:
cancer drug resistance database’, Sci Rep, vol. 3,
pp.1445.
Levatic, J, Curak, J, Kralj, M, Smuc, T, Osmak, M & Supek,
F 2013,’Accurate Models for P-gp Drug Recognition
induced from a cancer cell line cytotoxicity screen’, J
Med Chem, vol. 56, no. 14, pp. 5691-708.
Longley, DB & Johnston, PG 2005, ‘Molecular
mechanisms of drug resistanc’, J Pathol, vol. 205, no.
2,pp.275-92.
Ohta, Y, Yamamoto, Y, Okazaki, T, Uchiyama, T &
Takagi, T 1997, ‘Automatic Construction of
Knowledge base from Biological papers’, ISMB,-97
proceedings.
Pal, C, Palme, JB, Rensing, C, Kristiansson, E & Larsson,
J 2014, ‘BacMet: antibacterial biocide and metal
resistance genes database’, Nucleic Acids Res, vol. 42,
pp. D737–D743.
Proux, D, Rechenmann, F, Julliard, L, Pillet, V & Jacq, V
1998, ‘Detecting Gene Symbols and Names in
Biological Texts: A First step toward Pertinet
Information Extraction’, Genome Inform Ser Workshop
Genome Inform, vol. 9, pp. 72-80.
Remy, S, Gabriel, S, Urban, BW, Dietrich, D, Lehmann,
TN, Elger, CE, Heinemann, U & Beck, H 2003, ‘A
novel mechanism underlying drug resistance in chronic
epilepsy’, Ann Neurol, vol. 53, no. 4, pp.469-79.
Rindflesch, T, Tonabe, Lorraine, Weinstein, John &
Lawrence, H 2009, ‘EDGAR: Extraction of Drugs,
Genes and Relations from Biomedical Literature’, Pac
Symp Biocomput, pp. 517-528.
Walsh,C 2000, ‘Molecular Mechanisms that confer
antibacterial drug resistance’, Nature, vol. 406, no.
6797, pp.775-781.
Yeniterzi, S & Sezerman, U 2009, ‘EnzyMiner: automatic
identification of protein level mutations and their
impact on target enzymes from PubMed abstracts’,
BMC Bioinformatics, 10(suppl 8):S2.
ZK Drugresist - Automatic Extraction of Drug Resistance Mutations and Expression Level Changes from Medline Abstracts
173
