
Bridging the Gap between Knowledge Representation
and Electronic Health Records
Roberto Gatta
1
, Mauro Vallati
2
, Carlo Cappelli
3
, Berardino De Bari
4
, Massimo Salvetti
3
,
Silvio Finardi
5
, Maria Lorenza Muiesan
3
, Vincenzo Valentini
1
and Maurizio Castellano
3
1
Department of Radiation Oncology, Universit
`
a Cattolica del Sacro Cuore, Rome, Italy
2
School of Computing and Engineering, University of Huddersfield, Huddersfield, U.K.
3
University of Brescia, Dept. of Clinical and Experimental Sciences, Brescia, Italy
4
Radiation Oncology Department, Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland
5
A.O. Spedali Civili di Brescia, Brescia, Italy
Keywords:
Decision Support System, Computer Interpretable Guidelines, Electronic Health Record.
Abstract:
Decision Support Systems (DSSs) are systems that supports decision-making activities. Their application in
medical domain needs to face the critical issue of retrieving information from heterogeneous existing data
sources, such as Electronic Health Records (EHRs). It is well-known that there exists a huge problem of
standardisation. In fact, EHRs can represent the same knowledge in many different ways. It is evident that
the applicability of DSSs strongly relies on the availability of homogeneous collections of data. On the other
hand, the gap between DSSs and different EHRs can be bridged by exploiting middleware technologies.
In this paper, we tested CSL, a technology designed for working as a middleware between DSS and EHRs,
which is able to combine data taken from different EHR sources and to provide abstract and homogeneous
data to DSSs. Moreover, CSL has been used for implementing three Clinical Guidelines, in order to test its
capability in representing complex work-flows. The performed analysis highlight strengths and limitations of
the proposed approach.
1 INTRODUCTION
Decision Support Systems (DSS) are software sys-
tems designed to improve decision making. They are
widely exploited in a range of real-world domains,
such as business, military and economics. In the med-
ical domain, DSS are mainly used to support health-
care workers in one or more specific procedures, like
exams, investigation, diagnosis or therapies accord-
ing with the state-of-the-art of the specific domain of
practice. They are also commonly used for supporting
clinical research (i.e. clinical trials).
In order to provide support for a decision, a DSS
needs to match a reasoning work-flow with a set
of information take from available and relevant data
sources. Information can be provided manually by
operators but, usually, data are automatically taken
from existing Electronic Health Records (EHR): this
in order to limit errors and double data entry.
Consensus, Clinical Guidelines (CGs) and Inter-
nal Protocols and Procedures (IPP) are usually the
most common decision work-flows adopted in a clin-
ical environment. With the exception of the Con-
sensus, which normally are just a general agreement
about relevant clinical variables (and are not very de-
tailed from the procedural point of view), CG and
IPP are very similar and they describe the specific
best practice according to the clinical assessment of
the patient. Actually, they mainly differ from the le-
gal point of view: a physician tends to be more con-
strained to follow IPP rather than CG. This is because
IPP can be seen as a contextualisation of the more
general CG for the specific hospital. However, from a
mere Computer Science perspective, in the following
we will refer to both CG and IPP as Clinical Guide-
lines.
Clinical guidelines are usually written in natural
language with the exception of a few flow charts.
Since the knowledge codified in natural (and semi-
structured) language cannot be accurately exploited
by any existing automatic approach, a translation into
a formal language is required. The result of the trans-
lation, that comes into the form of an algorithm, is
usually referred to as Computer Interpretable Guide-
Gatta, R., Vallati, M., Cappelli, C., Bari, B., Salvetti, M., Finardi, S., Muiesan, M., Valentini, V. and Castellano, M.
Bridging the Gap between Knowledge Representation and Electronic Health Records.
DOI: 10.5220/0005648801590165
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 5: HEALTHINF, pages 159-165
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
159

line (CIG). A well-known formal language used for
encoding CIGs, also adopted by the HL7 consortium,
one of the major standard in the world of Medical
Informatics, is the Arden Syntax (Hripcsak, 1994):
it is based on the ideas of composing CG as a clin-
ical work-flow of several Medical Logic Modules
(MLMs). Arden Syntax, like other existing languages
designed for addressing similar issues (e.g., (Ohno-
Machado et al., 1998; Quaglini et al., 2001; Shahar
et al., 1998; Poikonen, 1997)), focuses on tasks that
need to be done. This in order to follow a pathway of
clinical procedure, and the control structures needed
to do it. A good comparison between existing lan-
guages is presented in (Peleg et al., 2003; Mulyar
et al., 2007).
Remarkably, in the context of the comparison it
has also been highlighted the problem of retrieving
existing patients from the different data sources. Due
to this reason, a complete system able to process CIGs
could need a large number of other technologies, in
order to get the required data stored in the different
data sources. Beside all the related issues, such as
differences in the granularity of stored information or
different encoding for the same sort of data, the adop-
tion of an intermediate software layer able to stan-
dardise the way for representing a specific clinical
meaning is required.
To standardize the knowledge representation in
the clinical domain, a major contribution (in partic-
ular for anatomical issues and their relationships, dis-
eases and relations between them or the anatomical
regions involved, etc..) comes from languages like
Resource Description Framework Schema (RDFS) or
Web Ontology Language (OWL). These approaches,
mainly derived from the work done on the Semantic
Web topic, have spawned the growth of many other
formalisms like query languages (e.g. SPARQL)
or languages for rule engines (e.g. OWL-S) (Ye
et al., 2009; Rold
´
an-Garc
´
ıa et al., 2009). However,
the aforementioned technologies are more oriented to
propose a way for representing the generic knowledge
of the medical domain rather than describe work-
flows in an efficient and compact manner.
At the state of the art, even though a number of
approaches have been proposed (Sordo et al., 2003;
Sujansky and Altman, 1996), retrieving data from ex-
isting EHRs in an automatic and affordable manner is
still considered a very complex task, due to the hetero-
geneity of EHR encodings. Moreover, existing EHRs
rely on different architectures in terms of Data Bases
(DBs), operating systems and servers. This represents
a critical barrier to the further diffusion of DSSs in
hospitals, because it forces the user to a double data
entry, which inevitably leads to mistakes and updating
issues, or to employ very expensive ad-hoc solutions.
One of the most interesting proposal is i2b2
1
but it
still has few applications and is more tailored to solve
problems of data acquisition and presentation for data
statistical analysis, rather than in the medical domain.
Nevertheless, i2b2 represents a promising first pro-
posal in an emerging scenario.
In this paper we focus on CSide Language (CSL).
The project CSide, now closed, aimed at building a
monolitic, advanced, multi-centric EHR. Specifically,
CSL has been thought as a language for representing
CIGs. Differently from existing similar languages,
CSL uses a different perspective. It sacrifices the ex-
pressivity of the language –from the point of view of
the clinical work-flows– in order to simplify the link
to available EHR and the real format of data. The
general idea of CSL relies on the fact that the most
interesting CGs are not those written between spe-
cialists of the same area, but guidelines written for
specialists of different areas (e.g., guidelines written
by cardiologists in order to be used by General Prac-
titioners). Evidently, due to the significant gap of
knowledge, the suggested work-flows are usually sim-
pler than those designed to be exploited by specialists
within the same topic, and the expressivity of the lan-
guage can therefore be reduced in order to make eas-
ier the access to the data.
In this paper we shortly introduce the CSL lan-
guage, and we propose a first experimental investi-
gation of such technology. Specifically, we tested
CSL on three different CGs, with a medium level of
complexity and very different structure (workflows vs
truth tables vs time role in the guidelines) in the real
clinical context of the EHR in a medium-size center
for studying Thyroid diseases. Moreover, we tested
CSL on a different EHR, in use in the Cardiovascu-
lar Diagnostic Center (CDC) to build, from available
data, more abstract concepts, i.e. the Circumferential
and Systolic Stress (cESS).
The remainder of this paper is organised as fol-
lows. Firstly, we describe the CSL architecture. We
then provide the results of the performed analysis. Fi-
nally, discussions and conclusions are given.
2 CSL
The architecture of CSL includes two layers, which
exploit different languages. The two level structure
has been designed for de-coupling EHR and DSS in-
terfaces. In CSL it is possible to modify the EHR
1
i2b2, “Informatics for Integrating Biology and the Bed-
side,” [Online]. Available: https://www.i2b2.org/. [Ac-
cessed 07 02 2014]
HEALTHINF 2016 - 9th International Conference on Health Informatics
160
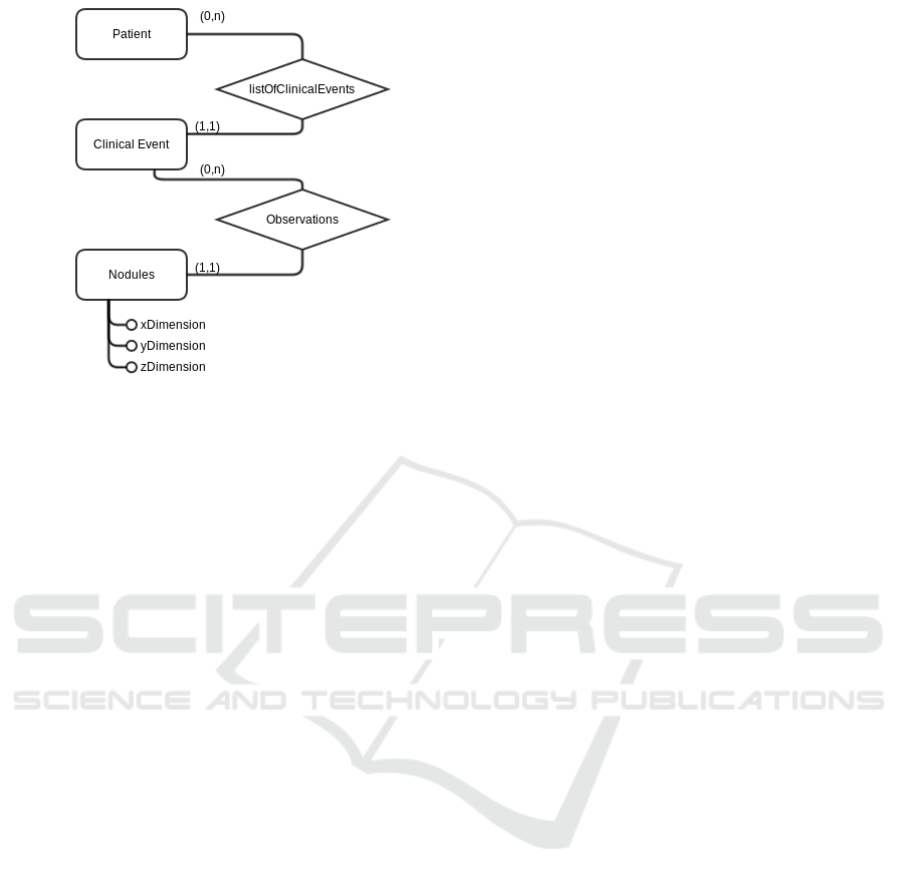
Figure 1: Example of a part of an ER schema.
or DSS structure (and interface) without changing all
the CSL code. The language used in the first level
is the Lower Level Language (LLL). It allows to cre-
ate, given the structure of database (tables), some en-
tities and relations between them, according with the
Entity Relationship (ER) paradigm. This level works
as a logical driver with respect to the data repository:
it hides the complexity of the EHR database schema,
and presents to the upper level more abstract and man-
ageable knowledge, that is useful for executing algo-
rithms and retrieving information. From this point of
view, it share some similarities with (Sujansky and
Altman, 1996) and an ORM (Object-Relational Map-
ping) tool.
The language exploited in the second layer is the
High Level Language (HLL) and it is used for build-
ing concepts based on the knowledge described in the
lower level. In order to implement an Object Oriented
(OO) paradigm, the HLL language inherits structure
and definitions from what has been defined in the
lower level using LLL. This can be done by inter-
preting the Entities as classes and the Relations as
methods. In the following sections we will present
a running example generated by using LLL and HLL
languages.
2.1 Lower Level Language
LLL uses simple primitives for building the entities,
and it works in a similar way as ORM languages does.
The definition of an entity can include one or more
tables and the relationships between them can be de-
fined using a SQL-like syntax. At runtime, the SQL-
queries are built automatically by the LLL engine.
Figure 1 shows part of an entity-relation schema.
This is actually a subset of the EHR used in the ex-
amined Thyroid Research Center of the Brescia City
Hospital. The translation of the two main entities
shown in Figure 1, namely Patient and Clinical Event,
and their relation, using the LLL language is the fol-
lowing.
define entity Patient based on table A01_PAT as
firstName as #IL{field:A01_NAME};
lastName as #IL{field:A01_LASTNAME};
birthDate as #IL{field:A01_BIRTHDATE};
IdPatient as #IL{field:A01_PATIENTID};
sex as #IL{field:A01_SEX};
firstName as #IL{field:A01_NAME};
with IdPatient as primary key;
with source testDB;
enddefine
define entity clinicalEvt base on
table A02_CLINICALEVENT as
IdClinicalEvent as #IL{field:A02_CLIEVID};
IdPatient as #IL{field:A01_PATIENTID};
fromDate as #{field:A02_FROMDATA};
toDate as #{field:A02_TO_DATE};
with IdClinicalEvent as primary key;
with source testDB;
enddefine
define left relation listOfClinicalEvent;
between Patient and clinicalEvt
on Patient.IdPatient and
clinicalEvt.IdPatient;
with source testDB;
enddefine
It should be noted that, in the provided LLL ex-
ample, also fields are defined. They are not shown in
Figure 1 for the sake of readability.
2.2 High Level Language
HLL has been designed with a strong focus on be-
ing “human readable”, even if it is a typed language.
There are two crucial points regarding HLL:
• a. Entities and relations defined in LLL are en-
coded, respectively, as classes and methods in
HLL. Methods are typically assigned to the mas-
ter entity in the case of a 1:n relation in LLL. By
exploiting this definition of classes, it is possible
to refer to an instance of an object by using the
Primary Key defined for the associated entity: the
CSL system automatically resolves the queries for
correctly handling the data on the Database.
• b. Using HLL it is possible to write complex algo-
rithms, called procedures, for managing methods
and attributes. The written procedure can then be-
come a new method for a chosen class. In this
way, starting from the few classes and methods
provided by LLL, through HLL it is possible to
Bridging the Gap between Knowledge Representation and Electronic Health Records
161

design and develop a more complex and abstract
set of methods. This allows to implement meth-
ods that do not suffer the high level of granularity
of the database, and that are closer to the abstract
semantic of the clinical domain.
An example of the HLL syntax is shown below. It
builds a method called ’BNAll’ for the class ’Patient’,
derived from the entity Patient previously described
in LLL, to calculate the biggest nodule’s dimension
measured in all the patients’ clinical events. To per-
form this task, it uses a method able of retrieving the
biggest dimension of the nodule described in a singu-
lar clinical event (the method ’BN’ working on class
’clinicalEvt’). It puts all the retrieved dimensions into
an array and returns the max value stored into the ar-
ray.
define procedure BNAll as method for entity
Patient on source testDB
set listToCheck = Patient.clinicalEvt
foreach listToCheck as PKclinEvent do
set nodDim = clinicalEvt(PKclinEvent).BN
put nodDim into array ArrayNodules;
endforeach
set values = Tools.getMaxArrayVal(diameters);
return(values);
enddefine
2.3 Features of CSL
For improving the usefulness of CSL, the following
features have been designed and implemented.
• Time: HLL offers several instructions to manage
temporal data and temporal relations, such as be-
tween, before and after.
• Domain: The domain can be applied both at the
LLL and at the HLL, and plays the same role of
the namespace in the weel-known XML language.
Specifically, it defines the scope where the enti-
ties, the relations or the procedures live.
• Type Declaration: HLL does not require an ex-
plicit type declaration for the variables. However,
HLL allows the developer to declare explicitly the
type, in order to avoid a wrong interpretation of its
content.
• Parameters: HLL can manage information from
various sources, and not only the considered
database. This is because, sometimes, not all the
relevant information are already stored within the
EHR; therefore, other input possibilities has to be
considered (external GUI, .csv file loading, ...). In
these situations HLL is able to receive input data
through command line parameters or other input
options.
3 METHODS
In this section we evaluate the CSL architecture us-
ing two real EHRs. The CSL has been used for im-
plementing three clinical guidelines regarding thyroid
diseases, and for building abstract data starting from
the available raw data.
3.1 The Testing Environment
A preliminary version of the CSL engine has been
made available. The engine for parsing and executing
scripts written in LLL and HLL has been developed
in PHP, and it exploits a MYSQL RDBMS for saving
concepts, entities, etc. This engine has been installed
on a Ubuntu 10.04-14+ server compiled with Sybase
driver and OCI driver, for giving the engine the ca-
pability of interact, respectively, also with SQLServer
and Oracle RDBMSs.
This preliminary experimental analysis is de-
signed for a first evaluation of the effective capa-
bility of CSL to be used in a real clinical context
for building useful and abstract concepts like CGs.
For evaluating the adaptability of CSL, we have in-
volved the Thyroid Center (CST) of the Brescia City
Hospital (Italy), which has recruited nowadays more
than 9,600 patients, and the Cardiovascular Diagnos-
tic Center (CDC), which has data for 12,500 patients.
For evaluating the capability of CSL to generate ab-
stract concepts, we implemented three CGs with a
similar level of complexity, but that exploit different
representation, using information from the CST. The
first CG is fully represented by a flowchart and heav-
ily involves temporal aspects; the second one is half
represented by a flowchart and half by a truth table,
and the third considered CG is only represented by a
truth table. The first and the second CGs chosen have
the same level of complexity of the CGs considered
in (Peleg et al., 2003).
Data from the CDC has been used to implement
two new abstract data; starting from the raw data
available in the EHR, we implemented the Circum-
ferential and Systolic Stress (cESS) abstract notion,
and notion of the changing of the percentage in vol-
ume of the ejection fraction between the first and
the last ultrasound examination. The CST database
includes more than 9,600 patients, and information
from more than 28,000 clinical events, 12,000 thyroid
ultrasound analysis and data describing more than
23,000 nodules are included; the CDC EHR includes
HEALTHINF 2016 - 9th International Conference on Health Informatics
162
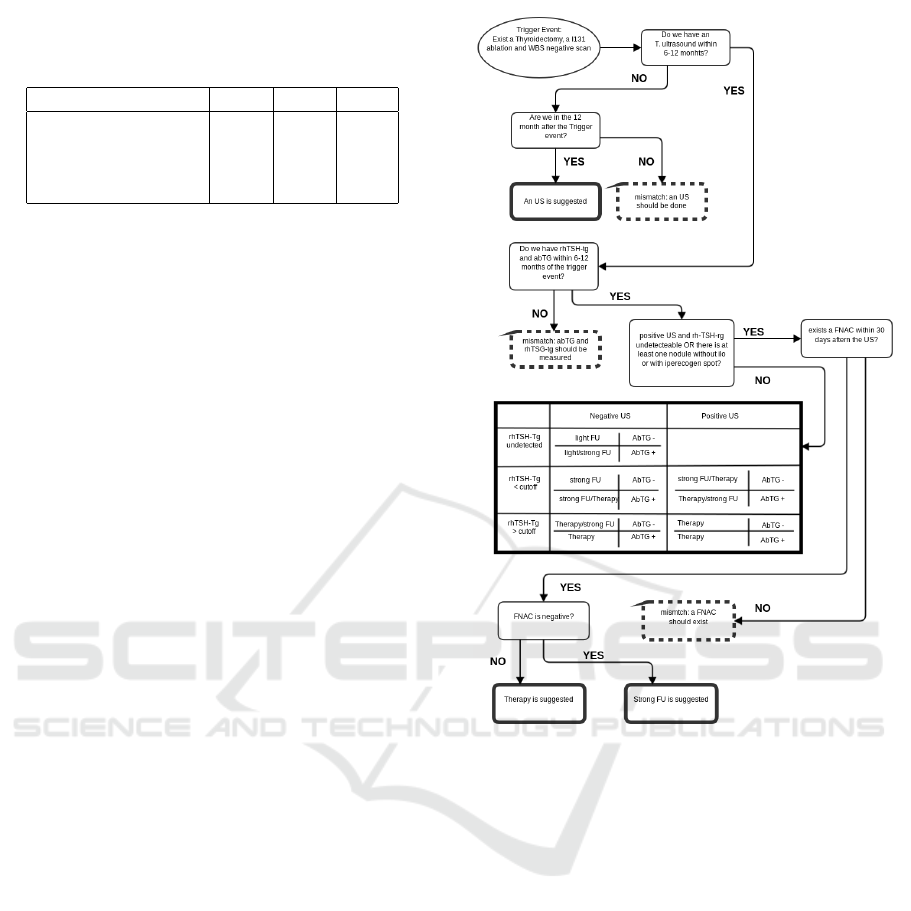
Table 1: The considered EHR described in terms of ar-
chitecture, number of recruited patients, number of clinical
events and other biometric measures.
CG1 CG2 CG3
Branches 29 6 0
Truth table 0 1 1
Exits 28 12 ?
Importance of Time High High Low
more than 12,500 patients and considers data from
around 16,000 cardiac ultrasound investigations.
When using EHRs form the Thyroid Center, ac-
cording to the requests of the physicians, we built the
entities and the concepts about the indication for Fine
Needle Aspiration Cytology (FNAC) under the suspi-
cious of thyroidal carcinoma, the patients are classi-
fied according to their pharmacological therapy and
on indications about the evolution of the follow up.
For evaluating the capability of CSL to implement
CIGs, we developed three procedures to check if a
patients clinical pathway agrees with three different
guidelines. Specifically:
• CG1. These CGs evaluate if a patient should re-
ceive a Fine Needle Aspiration Cytology proce-
dure to rule out thyroidal carcinoma (Pacini et al.,
2006);
• CG2: evaluate if follow up after 131I and/or
surgery treatment as indicated (Pacini et al.,
2006);
• CG3: suggest ablation versus surgical therapy for
the management of Thyroid nodules and Differ-
entiated Thyroid Cancer (Cooper et al., 2009).
Before the implementation in CSL, the three CGs
were translated in a semi-structured way (a flow-chart
and/or a truth table) as described in the following ta-
ble. Figure 2 shows the mixed data-flow/truth table
structure of CG2.
The data from the Cardiovascular Diagnostic Cen-
ter has been used for generating two more manageable
and abstract notions. In fact, their implementation us-
ing CSL was straightforward. This is because both of
the concepts derives from quite simple computations
that do not consider (or consider a very limited set
of) temporal aspects; the construct provided by CSL
made quite trivial their representation and computa-
tion. Nevertheless, CSL is capable of effectively sup-
port the implementation of such sort of notions, too.
3.2 Discussion
This section is devoted to discuss results and inter-
esting observations about the use of the CSL in our
testing scenarios.
Figure 2: Data-flow/truth table of the CG2 (Pacini et al.,
2006). With bold-continuous borders, the suggested next
actions, in bold-dotted borders, the detected mismatch. In
this CG time plays an important role, and the single con-
cepts in the steps can be strongly abstracted with respect to
the granularity of EHR data.
3.2.1 CST
The process of encoding the Clinical Guidelines into
CIGs has been divided into two steps: firstly, CGs
were translated in a semi-structured language (i.e.
flowchart or truth table), then the semi-structured CG
was translated in CSL structure. We noticed that the
first step was the most complex. Even if the CG was
well-known and commonly adopted, different spe-
cialists suggested slightly different flowcharts due to
the high level of ambiguity of the original guideline.
This would suggest to put more attention in writing
such CGs because a large part of knowledge is not ex-
plicitly given, but considered as implicitly known, and
the temporal aspects are often not considered (e.g., for
how long a AbTG exam could be considered valid,
and when should it be considered too obsolete for the
Bridging the Gap between Knowledge Representation and Electronic Health Records
163

computation?).
In general, the process of coding in CLS was quite
simple and, with the exception of few bugs due to
the prototype of the engine, it was immediately ex-
ploitable. We also observed that the code is easy to
read and understand: this is a pivotal element for al-
lowing and effective maintenance of the system. Nev-
ertheless, it is clear that CSL is still in a very prelim-
inary shape, and many improvements would be sug-
gested: for example a Graphical User Interface would
be very helpful for managing concepts. Also, the link
to the database it is not easy to be set and the domains
part should be better implemented.
We also identified potential improvements on the
time handling side of CSL even if, in general, the
available constructs was enough to solve our prob-
lems. A major improvement is required in the interac-
tion between CSL and other IT infrastructure, a point
that in the prototype is very weak because it forces
the user to program in PHP and eventually to write a
wrapper function to make CSL to be able to interact
via web services: we think that this interoperability
should be provided directly to the engine.
According to the observed results and to the ex-
perience gained, we believe that the proposed archi-
tecture can have very practical applications in com-
mercial EHRs. This is mainly due to the fact that
CSL merges the most diffused modelling and design
paradigms (ER schema, OOP) with the simplicity of
the script language. Moreover –but this will require
some language and engine improvements– CSL could
give an interesting contribution to face the well known
“curly braces” problem. However, this goal also re-
quires an industry-wide standardisation. Therefore,
this direction can be explored in association with ven-
dors, in order to allow a larger analysis.
3.2.2 CDC
CSL made the process of describing the abstract data
notions very easy and smooth. According to received
feedback, it is suitable to be used also for such pur-
poses. From this point of view, CSL could really
represent a valid middleware between the RDBMs
and more specialised software like DSSs and or CIGs
process environments. Nevertheless, further improve-
ments in the interaction process with other IT archi-
tecture should be provided and the engine should be
made more stable.
4 CONCLUSION
DSS are effectively and widely exploit in many do-
mains, but in the medical domain their diffusion and
exploitation is slowed by the very different technolo-
gies used by existing EHRs. In this paper we de-
scribed and tested CSL, a middleware system for
bridging the gap between DSSs and data sources in
the medical domain.
The proposed technology has been tested by using
data and involving experts of the Thyroid Research
Center and the Cardiovascular Diagnostic Center of
the Brescia City Hospital. The Thyroid Research
Center, in particular, has a complex EHR architecture
and different CIGs has been implemented: in this real
world environment CSL has shown a good aptitude
for handling data from SQL-like databases in order
to build, according with the knowledge expressed by
HLL scripts, abstract concepts and present to the up-
per level (i.e. to a DSS) an homogeneous view of the
data stored in the databases. CSL demonstrated to be
able to represent CIGs and to perform complex rea-
soning processes on the retrieved data. Despite this,
we still suggest to consider CSL as a technology for
retrieving data from different EHRs and presenting
them to DSSs; more tests are needed before consid-
ering CSL as a language for developing DSSs.
Future work include a thorough investigation of
the capabilities of CSL for handling complex reason-
ing on the extracted data, as well as a larger experi-
mental analysis on EHRs from different medical de-
partments. Finally, we plan to further improve the
usability of the CSL by exploiting feedback received
from medical experts.
REFERENCES
Cooper, D. S., Doherty, G. M., Haugen, B. R., Kloos, R. T.,
Lee, S. L., Mandel, S. J., Mazzaferri, E. L., McIver,
B., Pacini, F., Schlumberger, M., Sherman, S. I., Stew-
ard, D. L., and Tuttle, R. M. (2009). American thyroid
association (ata) guidelines taskforce on thyroid nod-
ules and differentiated thyroid cancer, revised amer-
ican thyroid association management guidelines for
patients with thyroid nodules and differentiated thy-
roid cancer. Thyroid, 19(11):1167–1214.
Hripcsak, G. (1994). Writing arden syntax medical
logic modules. Computers in biology and medicine,
24(5):331–363.
Mulyar, N., vanderAalst, W. M., and Peleg, M.
(2007). A pattern-based analysis of clinical computer-
interpretable guideline modeling languages. J Am
Med Inform Assoc, 14(6):781–787.
Ohno-Machado, L., Gennari, J. H., Murphy, S. N., Jain,
N. L., Tu, S. W., Oliver, D. E., Pattison-Gordon,
HEALTHINF 2016 - 9th International Conference on Health Informatics
164

E., Greenes, P. A., and E. H. Shortliffe, G. O. B.
(1998). The guideline interchange format: a model
for representing guidelines. J Am Med Inform Assoc.,
5(4):357–372.
Pacini, F., Schlumberger, M., Dralle, H., Elisei, R., Smit,
J. W., and Wiersinga, W. (2006). European consen-
sus for the management of patients with differenti-
ated thyroid carcinoma of the follicular epithelium.
european thyroid cancer taskforce. Eur J Endocrinol,
154(6):787–803.
Peleg, M., Tu, S., Bury, J., Ciccarese, P., Fox, J., Greenes,
R. A., Hall, R., Johnson, P. D., Jones, N., Kumar,
A., Miksch, S., Quaglini, S., Seyfang, A., Short-
liffe, E. H., and Stefanelli, M. (2003). Compar-
ing computer-interpretable guideline models: a case-
study approach. J Am Med Inform Assoc, 10(1):52–
68.
Poikonen, J. (1997). Arden syntax: the emerging standard
language for representing medical knowledge in com-
puter systems. Am J Health Syst Pharm, 54(3):281–
284.
Quaglini, S., Stefanelli, M., Lanzola, G., Caporusso, V., and
Panzarasa, S. (2001). Flexible guideline-based patient
careflow systems. Artif Intell Med, 22(1):65–80.
Rold
´
an-Garc
´
ıa, M., Navas-Delgado, I., Kerzazi, A.,
Chniber, O., Molina-Castro, J., and Aldana-Montes,
J. F. (2009). Ka-sb: from data integration to large scale
reasoning. BMC Bioinformatics, 1(10):10–S5.
Shahar, Y., Miksch, S., and Johnson, P. (1998). The asgaard
project: a task-specific framework for the application
and critiquing of time-oriented clinical guidelines. Ar-
tif Intell Med, 14(1-2):29–51.
Sordo, M., Ogunyemi, O., Boxwala, A. A., and Greenes,
R. A. (2003). Gello: an object-oriented query and ex-
pression language for clinical decision support. AMIA
Annu Symp Proc, page 1012.
Sujansky, W. and Altman, R. (1996). An evaluation of the
transfer model for sharing clinical decision-support
applications. Proc AMIA Annu Fall Symp, pages 498–
472, Washington, D.C., Hanley & Belfus.
Ye, Y., Jiang, Z., Diao, X., Yang, D., and Du, G. (2009). An
ontology-based hierarchical semantic modeling ap-
proach to clinical pathway workflows. Comput Biol
Med, 39(8):722–732.
Bridging the Gap between Knowledge Representation and Electronic Health Records
165
