
POMap: An Effective Pairwise Ontology Matching System
A. Laadhar
1
, F. Ghozzi
2
, I. Megdiche
1
, F. Ravat
1
, O. Teste
1
and F. Gargouri
2
1
University of Toulouse, IRIT (CNRS/UMR 5505), 118 Route de Narbonne, 31062 Toulouse, France
2
University of Sfax, MIRACL, Sakiet Ezzit 3021, Tunisia
Keywords:
Semantic Web, Ontology Matching System, Syntactic Matching, Structural Matching.
Abstract:
The identification of alignments between heterogeneous ontologies is one of the main research issues in the
semantic web. The manual matching of the ontologies is a complex, time consuming and an error prone task.
Therefore, ontology matching systems aims to automate this process. Usually, these systems perform the
matching process by combining element and structural level matchers. Selecting the optimal string similarity
measure associated with its threshold is an important issue in order to enhance the effectiveness of the element
level matcher, which in turn will improve the whole ontology system results. In this paper, we present POMap,
an ontology matching system based on a syntactic study covering element and structural levels. For the element
level matcher we have adopted the best configuration based on the analysis of the performances of many string
similarity measures associated with their thresholds. For the structural level, we have performed a syntactic
study on both subclasses and siblings in order to infer the structural similarity. Our proposed matching system
is validated and evaluated on the Anatomy, the Conference and the Large Biomedical tracks provided by the
benchmark of OAEI 2016 ontology matching campaign.
1 INTRODUCTION
To ensure the interoperability between different plat-
forms, ontologies has been used as the essence of the
semantic web. An ontology can model a particular
domain as well as the relationships between its enti-
ties in order to allow data to be shared, reused, inte-
grated and queried by different stakeholders. Ontol-
ogy matching is the process of finding a set of map-
pings between the entities of two or more ontologies
representing a similar domain (Shvaiko and Euzenat,
2013). Therefore, the ontology matching community
has been proposing a variety of strategies in order to
automate the ontology matching process.The ontol-
ogy matching process takes as an input two ontolo-
gies, which contain a set of entities. Usually the map-
pings between two entities is evaluated by a score
of similarity ∈ [0,1]. This score defines the degree
of confidence of a mapping relationship. This confi-
dence value is configured automatically by the match-
ing system. A matcher is an algorithm that processes
two ontologies and returns the set of mappings be-
tween them along with a similarity value. Usually,
a matching system combines several distinct match-
ers depending on the input ontologies characteristics.
This combination aims to improve the final align-
ment effectiveness. A single matcher is not sufficient
to achieve a high-quality alignments results. Match-
ers are classified into two main levels: element level
matchers and structure level matchers (Shvaiko and
Euzenat, 2013). Each level can rely on different tech-
niques and methodologies such as syntactic, semantic
and structural based matching.
In this paper, we address the problematic of on-
tology matching while focusing on the syntactic char-
acteristics of both element level and structural level.
As a first step, we focus on finding the optimal sim-
ilarity measure and threshold in order to perform the
element level matching process. The two target on-
tologies are matched using the element matcher while
exploring all the names of an ontology class, such as
labels, local name and synonyms. We performed a
series of tests in order to define the optimal similar-
ity measure and threshold. Since we are adopting a
sequential composition, the output of element level
matcher is the input of the structural based matcher.
Dealing with the structural matching, Almost the ex-
isting research works focus on the propagation of the
syntactic similarity score in order to derive new struc-
tural mappings. However, this propagation do not
take into consideration the syntactic characteristics of
the target entities. Therefore, we propose two struc-
Laadhar A., Ghozzi F., Megdiche I., Ravat F., Teste O. and Gargouri F.
POMap: An Effective Pairwise Ontology Matching System.
DOI: 10.5220/0006492201610168
In Proceedings of the 9th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (KEOD 2017), pages 161-168
ISBN: 978-989-758-272-1
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

tural matchers, guided by the syntactic treatments on
the subclasses and the sibling entities without mea-
suring any structural similarity score. Each structural
matcher focuses on a part of the ontology hierarchy in
order to obtain a new set of alignments. These match-
ers take profit of the hierarchical relationships of an
ontology siblings and subclasses exploiting by a syn-
tactic treatments of common words.
The remainder of this paper is organized as fol-
lows: Section 2 discusses the existing ontology
matching systems. Section 3 presents the POMap
ontology system including the element and structural
level matchers. Section 4 evaluates and discusses the
obtained results. Conclusion and suggestions for fu-
ture work are shown in section 5.
2 RELATED WORKS
Matching Levels. Usually, a matching system em-
ploys different matchers which in turn follow dis-
tinct matching techniques (Otero-Cerdeira et al.,
2015)(Megdiche et al., 2016).
The element level matchers determine the map-
pings between two entities without taking into con-
sideration the existing relationships between these en-
tities. The state of the art element level matchers
are mostly relying on string based techniques, which
compute the similarity between two entities through
syntactic similarity measures. Language-based tech-
niques can also be employed by the element level
matchers. These techniques rely on natural language
processing (NLP) such as the stop words removal and
lemmatisation in order to derive meaningful terms.
Constraint based techniques can be also employed in
order to analyse the internal structure of an entity like
the domain, range as well as the cardinality of the at-
tributes.
Structure level matchers can use external knowl-
edge while taking profit of the relationships between
the entities (classes, instances and properties). Re-
searchers have been employing several techniques
such as graph-based and instance based. The graph
based techniques compute structural similarities be-
tween neighbouring entities based on their position
in the ontology. Following this intuition, Similarity
Flooding (SF) (Melnik et al., 2002) is a well-known
structural matcher inspired by the idea of structural
similarity propagation. This technique propagates the
structural similarity score among the ontology candi-
date mappings. In another research work, (Cruz and
Sunna, 2008) proposed two structural matchers: De-
scendants Similarity Inheritance (DSI) and Siblings
Similarity Contribution (SSC). For a given set of ini-
tial mappings discovered by an element level matcher,
the main idea of DSI and SSC is to refine the initial
element level mappings with weighted scores accord-
ing to the descendants and the siblings. DSI slightly
outperform the Similarity Flooding method. How-
ever, even after applying these structural matchers,
the value of F-measure still not good enough in terms
of effectiveness. We argue that this inefficiency is
caused by the bad quality of the initial alignments re-
sulted our first matcher. In this research work, dif-
ferentiated from the state of the art, we do not mea-
sure any structural similarity score. Therefore, we are
guided by the initial similarity score in order to de-
rive new structural mappings while taking the rela-
tionships between the entities as a discriminating fea-
ture. These initial mappings are the input of the struc-
tural level matcher in order to deduce a new set of
mappings.
Ontology Matching Systems. We review some of
the top relevant and mature matching systems, which
had a significant success during the last years of the
OAEI campaign. Our results will be compared with
these matching systems on section 4.
Agreement Maker Light (AML) (Faria et al.,
2013) is an automated large-scale ontology matching
system focusing on the biomedical ontologies match-
ing. AML translates the entities of the input ontolo-
gies into the same language. Then, AML employs
several matchers such as a lexical matcher, an external
background knowledge matcher, a structural matcher
and an instance matcher. AML follows a sequential
composition. The first matcher implemented by AML
is the Lexicon Matcher, which searches for exact
matching between the entities names of the two input
ontologies. AML employs also a mediating matcher,
which take profit of some external knowledge sources
in order to enrich the input ontologies by new tex-
tual content. In the OAEI 2016 compaign, AML used
the following external knowledge resources: Uber
Anatomy Ontology (Uberon) (Mungall et al., 2012),
the Human Disease Ontology (DOID) (Schriml et al.,
2011), the Medical Subject Headings (MeSH) (Nel-
son et al., 2001) and Wordnet (Miller, 1995). AML
derives its structural matcher from the DSI algorithm.
LogMap (Jim
´
enez-Ruiz and Grau, 2011) is an on-
tology matching system focusing on both the ele-
ment level and the structural level matching of large
biomedical ontologies. The architecture of LogMap
is divided into two main stages: the computation of
candidate mappings and their assessment. In the first
stage, LogMap extracts a set of initial candidate map-
pings using a lexical comparison method. These can-
didate mappings are extracted from the inverted index
of each input ontology. An inverted index contains
the labels and the URIs of each ontology. In the sec-
ond stage, LogMap tries to remove the set of incor-
rect candidate mappings by applying lexical, struc-
tural and reasoning techniques.
XMap (Djeddi and Khadir, 2014) deals especially
with the problem of scalability. XMap follows a
sequential composition of three layers: terminolog-
ical Layer, structural layer and alignment layer. In
the terminological layer, XMap employs a transla-
tion matcher in case of multilingual ontologies. Also,
XMap uses a string matcher, which computes the sim-
ilarity between the textual descriptions of two en-
tities by applying some semantic measures. Xmap
employs two background knowledge sources word-
net (Miller, 1995) and UMLS (Bodenreider, 2004)).
During the structural layer, XMap computes the sim-
ilarity between two entities by taking into account
the internal structure of a concept. Finally, in the
alignment Layer, the set similarity scores resulted
by the string matcher, the linguistic matcher and the
structural-based matcher are aggregated using mathe-
matical methods.
CroMatcher (Guli
´
c et al., 2016) is an ontology
machting system concentrating on the aggregation
of different matchers using a weighted factor. Cro-
Matcher divides its matchers into two main sets:
string based and structure based. The string based
matchers determines the mappings between two en-
tities relying on several techniques such as syntac-
tic similarity measure, instance matching and prop-
erty matching. CroMatcher performs the structural
matching using four distinct matchers: Super En-
tity matcher, sub entity matcher, domain matcher and
range matcher. The matching system aggregates the
results of the string matchers and the structural match-
ers by automatically giving a weight for each sin-
gle matcher. The two obtained matrices of the string
based matcher and structural based matcher are also
combined using an automated weighting factor in or-
der to obtain the final set of alignments.
3 THE POMAP ONTOLOGY
MATCHING SYSTEM
3.1 POMap Matching System Overview
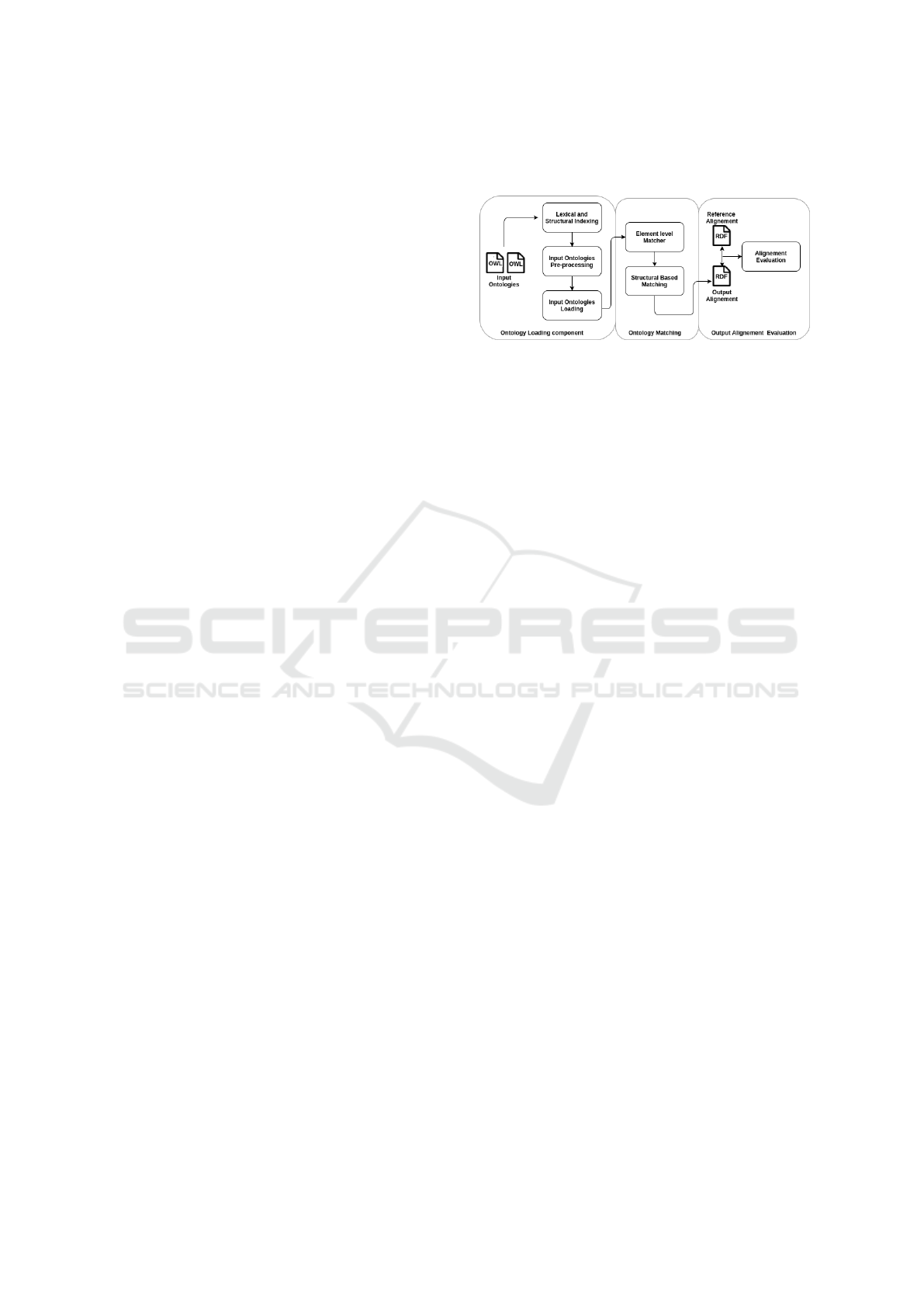
The workflow of POMap is depicted in Figure 1.
This workflow contains several steps beginning by the
OWL ontologies loading until the RDF output align-
ment evaluation. The architecture of POMap contains
three main components: The ontology loading, the
ontology matching and the output alignment evalua-
tion. We will describe in the following sub-sections
the set of activities performed by each component.
Figure 1: The architecture of POMap.
3.2 Ontology Loading Component
We parse a given set of two input ontologies using
OWL API in order to populate our lexical and struc-
tural data structure. The lexical data structure is a
multimap that contains the triplet: a set of classes,
their names as well as the type of each names. A
name has one of these types: URIs, labels or syn-
onym properties. In terms of the structural data struc-
ture, we store all the relationships between classes. A
pre-processing techniques are mandatory to acquire
a cleaner data-set. Since several similarity measures
take into consideration the letter case as a parame-
ter influencing the similarity score, we set in lower-
case all the input ontology names. Next, we discard
all stopwords as well as the non-alphanumeric char-
acters. We also replace every underscore to a space
character. Finally, we perform an English stemming
process as the one proposed by Porter (Porter, 2001).
3.3 Element Level Matcher
In order to discover mappings, ontology match-
ing systems employ one or more similarity mea-
sures without any reference to the defined thresholds.
Therefore, we tested the different similarity measures
in order to define the optimal measure and threshold
after performing the pre-processing steps. After the
pre-processing steps, the input ontologies are quite
ready to be processed by our element level matcher.
This matcher is responsible for computing the simi-
larity score between all the class names of the first
and the second ontology. To perform that, we fol-
low an all-against-all strategy, which stands for mea-
suring the similarity score between all the the pos-
sible combinations of the entities of the two ontolo-
gies. Then, we choose the maximum similarity score
for every pair of resulted candidate mappings. We
argue that comparing all the names of the two in-
put ontologies is more efficient than comparing only
the labels. For instance, our element level matcher

achieved an F-Measure of 0.830 using only the la-
bels on the anatomy OAEI dataset. However, using
all the names associated with the Anatomy entities,
we succeeded to accomplish an F-Measure of 0.862.
Therefore, we are able to better capture the syntactic
mappings between two ontologies and achieve better
results. We employed similarity measures in order to
compare pairwise names. The efficiency of a syntac-
tic matcher is determined by the trade-off between its
resulted F-measure value and its runtime. For exam-
ple, a non-efficient syntactic matcher can produce a
high F-measure in a very long time. The variety of
syntactic similarity measures arises the difficulty of
choosing the optimal one associated with its thresh-
old. The optimal choice of the similarity measure and
threshold will immediately improve the alignment re-
sults especially in terms of F-measure. Some re-
searchers (Duan et al., 2010) (Cheatham and Hitzler,
2013) (Ngo et al., 2013) (Sun et al., 2015) analysed
the performance of similarity measures while dealing
with the ontology matching process. However, none
of them analysed the performance of these measures
after performing the pre-processing based on all the
names of an ontology class. Moreover, almost of re-
searchers did not included ISUB (Stoilos et al., 2005)
in their comparison. Consequently, we will reveal the
best similarity measure in terms of F-measure, thresh-
old and runtime. In order to select the optimal F-
Measure, we apply a trimming process using a differ-
ent threshold value for each syntactic similarity mea-
sure. As shown in figure 2, for every experiment, the
element level matcher has as an input a single mea-
sure associated with a threshold value. The output of
the testing process are the set of evaluation criteria:
the recall, the precision and the F-Measure. During
our series of tests, we vary the threshold value from
the range of 0.1 to 1 with an interval of 0.01.
Figure 2: The input/output schema of the element level test-
ing process.
Then, we compute the F-measure for each thresh-
old interval. The employed tracks in our testing pro-
cess are: the Anatomy track and the Large Biomed-
ical track. For the later, we evaluated the avail-
able similarity measures used by our element level
matcher through the three small fragments: FMA-
NCI , SNOMED-FMA and SNOMED-NCI. Figure 3
draws the variation of F-Measure while adopting dif-
ferent thresholds in the Anatomy track of OAEI. Fig-
ure 4, Figure 5, Figure 6 and Figure 7 show the value
of the optimal F-measure and threshold obtained by
each similarity measure for each OAEI track. The
other tested state of the art syntactic similarity mea-
sures are available at: https://goo.gl/1kUgkH. On all
the three tasks, ISUB with a threshold of 0.9 outper-
form the other similarity measures. We conclude that
ISUB 0.9 is the best measure for ontologies syntactic
matching associated with the pre-processing steps. To
optimise the execution time we took profit of the multi
core architecture of the processor. Therefore, we
adopted an intra-matching parallelism methodology.
Thus, the element level matcher is divided into the
number of available CPU cores and executed in paral-
lel. For a given two input ontologies, the algorithm of
the element level matcher loops classes names of the
source and target indexes and derives for each class
index the set of corresponding names. These names
are compared to each other using a syntactic simi-
larity measure (ISUB). Only the pair of names hav-
ing a threshold above the 0.9 are added to the set of
mappings. Since we are pursuing an alignment multi-
plicity of 1:1, we followed the element level matcher
by a cleaning process of the resulted candidate map-
pings. Therefore, we keep only the maximum similar-
ity score that a single class can have. If two mappings
have the same similarity score we select randomly one
of them.
3.4 Structural Level Matching
For a given set of alignments, which are discovered
by the element level matcher, we can enrich these
mappings by a set of new ones by applying the struc-
tural matching. Our structural level matcher is com-
posed of two structural sub-matchers: siblings and
subclasses. During the first sub-matcher, we follow
the intuition that states: if two classes are similar, then
their siblings should be somehow similar (Shvaiko
and Euzenat, 2013). In the second sub-matcher, we
remove the common words between the super class
and its subclasses. Moreover, this matcher admit the
idea that: if two classes are similar, then their sub-
classes should be also similar. In that way, the struc-
tural matching improves the initial matching results
of the element level matcher.
3.4.1 Structural Matcher based on Siblings
This structural matcher detects new mapping based
on the siblings of the already discovered alignments
by the element level matcher. These mappings have
a similarity score between ISUB 0.9 and ISUB 0.8.
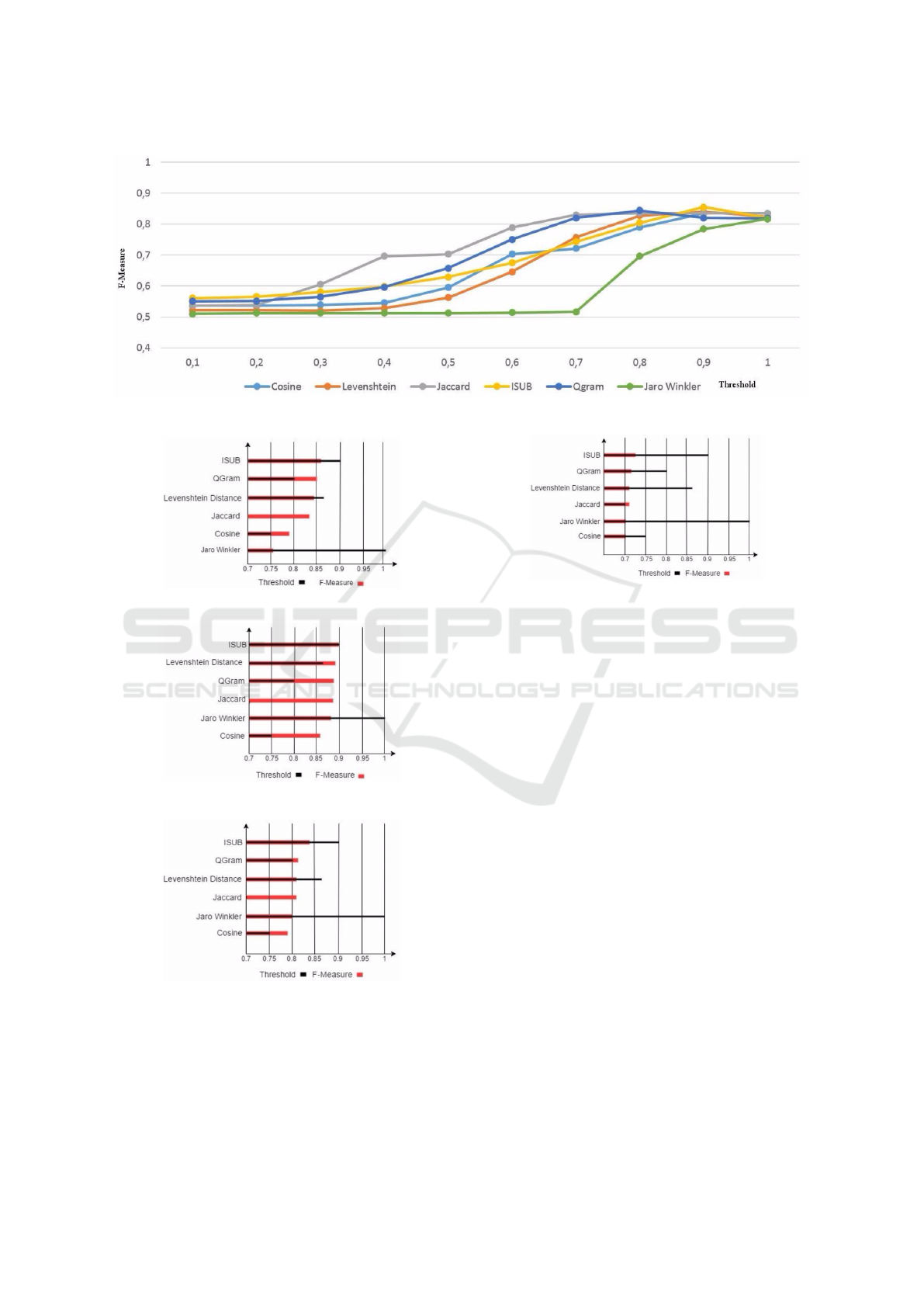
Figure 3: The variation of F-Measure over the Anatomy track while adapting different similarity measure and thresholds.
Figure 4: Top F-Measure in Anatomy task.
Figure 5: Top F-Measure in FMA-NCI small fragment.
Figure 6: Top F-Measure in FMA-SNOMED small frag-
ment.
We argue that if two entities are matching, then their
siblings will probably match. Therefore, we decrease
the threshold from 0.9 to 0.8. For a given ontology O
and an entity e
O
, the j
th
sibling of an entity e is de-
noted as Sibj
O
e
, where
∑
Sibj
O
e
∈ [0,m] and m is the
Figure 7: Top F-Measure in SNOMED-NCI small frag-
ment.
number of siblings. We consider a single mapping re-
sulted by the element level matcher as: a
i
= (e
O
k
,e
O
0
L
,s)
with Sib1
O
ek
is the only sibling of e
O
l
. If the similar-
ity score between the two siblings is high (still below
the syntactic threshold), then we consider that the two
siblings will certainly match. However, if the two en-
tities e
O
K
and e
O
0
L
have many siblings, then we should
compare all the siblings of e
O
k
with the siblings of e
O
0
l
.
Therefore, we perform an all-against-all comparison
between all the siblings of e
O
k
and e
O
0
l
. Next, like the
element level matcher, we pursue an alignment mul-
tiplicity of 1:1. The input of the sibling matching al-
gorithm is the discovered mappings by the syntactic
matcher as well as the two input ontologies. The out-
put of this matcher is the new set of structural map-
pings, which are added to the initial mappings dis-
covered by the element level matcher. Since we used
a threshold of 0.8 ISUB for the structural matcher, we
removed all the mappings, which are already discov-
ered by the syntactic matcher.
3.4.2 Structural Matcher based on Subclasses
For the subclasses structural matcher, we are follow-
ing the intuition that if two classes are similar, then
their sub classes are similar (Shvaiko and Euzenat,
2013). Given an ontology O and an entity e of O,
the j
th
descendant of e
O
is denoted as Subj
O
e
, where
∑
Sub
O
e
∈ [0,n] and n is the number of descendants. To
illustrate, if a single mapping is denoted as a
i
= ( e
O
k
,
e
O
0
L
, s), Subj
O
ek
is the descendent of e
O
k
and Subj
O
0
el
is
the descendent of e
O
0
l
. Therefore, Subj
O
0
el
and Subj
O
ek
are probably similar. However, if there are many sub-
classes of e
O
k
and e
O
0
l
, it will be complex to perform
the mapping process between the descendant, espe-
cially when they have close similarity scores. There-
fore, it is necessary to find some other discriminating
features (Shvaiko and Euzenat, 2013). To illustrate,
we consider an example of mapping resulted from the
element level matcher: a
i
= ( e
O
k
, e
O
0
l
, s), with s >
threshold and the name of e
O
k
= e
O
0
l
= ”heart ventrice”.
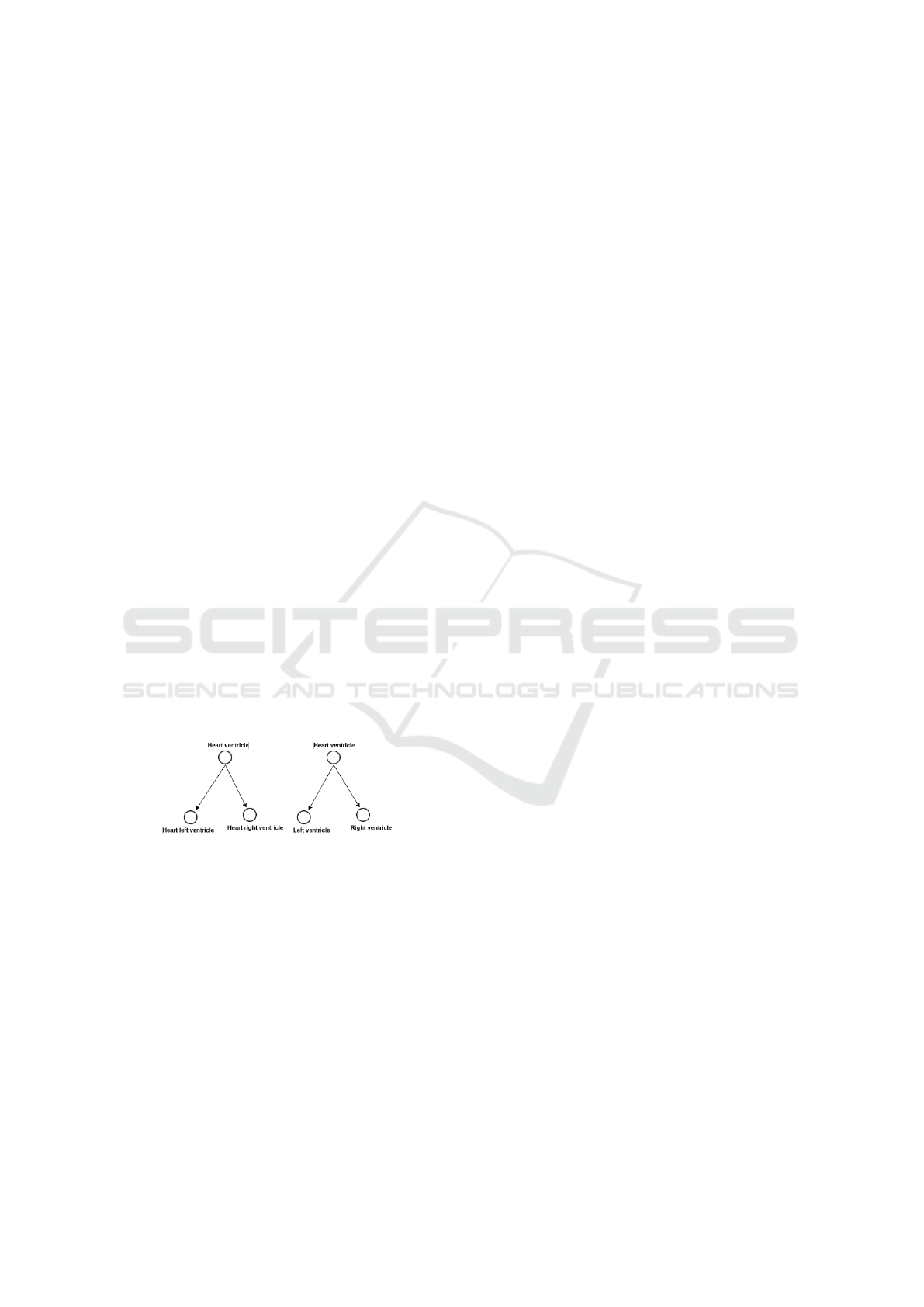
For simplicity, as shown in the figure 8, each entity of
this mapping has only two subclasses:
• Sub1
O
ek
, Sub2
O
ek
where the name of Sub1
O
ek
= heart
left ventricle and the name of Sub2
O
ek
= ”heart
right ventricle”.
• Sub1
O
0
el
, Sub2
O
0
el
, where the name of Sub1
O
0
el
=
”left ventricle” and the name of Sub1
O
0
el
= ”right
ventricle”
Then, we remove the common tokens between the
entity e
k
O
and its descendants (Sub1
ek
O
, Sub2
ek
O
).
We also remove the common tokens between e
l
O
0
and
its descendants (Sub1
el
O
0
, Sub2
el
O
0
) . Consequently,
the descendants (sub-classes) became:
• Sub1
O
ek
= ”left” and Sub2
O
ek
= ”right”
• Sub1
O
0
el
= ”left” and Sub2
O
0
el
= ”right”
Figure 8: Structural matching based on subclasses illustrat-
ing example.
As a result, we can match the descendants of
e
k
O
( Sub1
O
ek
, Sub2
O
ek
) and the descendants of
e
O
0
l
(Sub1
O
0
el
, Sub2
O
0
el
) by computing the similarity
score between them (all-against-all strategy). Since
we are following a 1:1 multiplicity, then we keep
only the maximum score for each class of the candi-
date structural mappings, which has a similarity score
above the threshold (Monge Elkan 0.85). Unlike
ISUB, Monge Elkan (Monge et al., 1996) is able to
capture the mappings between two textual sequences,
which are containing numerical values. Furthermore,
Monge Elkan is not recommended for large matching
tasks, as a result we are using it only for the structural
matching process. The new discovered mappings are
added to the list of initial alignments which are identi-
fied by the element level matcher. The mappings data
structure contains the list of the already discovered
mappings from the two input ontologies. We notice
that we pursued a cleaning process of the discovered
mappings by the structural matcher in order to dis-
card the list of mappings already found by the element
level matcher as well as structural matcher based on
siblings.
4 EXPERIMENTS AND
DISCUSSIONS
4.1 Experimental Setup
We developed the POMap system using JAVA and
Eclipse as the integrated development environment.
We choose to perform the experiment process analy-
sis through two biomedical tracks of the OAEI bench-
mark as well as the conference track in order to show
the efficiency of POMap in various matching tasks
while dealing with OWL ontologies. We employed
for our series of tests a hardware configuration of:
15Gb of RAM coupled with an Intel Core i5-4570
CPU @3.40 Ghz x 4. This configuration is similar
to the one used by the OAEI benchmark. We com-
pare the performance of POMap with the results of
ontology matching system enrolled in the OAEI 2016
campaign obtained using the Anatomy, Conference as
well as the LargeBio track.
4.2 Experimental Results
Anatomy. The anatomy track is a matching task
between the Adult Mouse Anatomy (2744 classes)
and the NCI Thesaurus (3304 classes) describing the
human anatomy. In terms of F-Measure, POMap
is ranked in the fourth position among 14 match-
ing systems while producing a competitive runtime
of 43 seconds. We achieved an F-Measure of 0.893
without using any external knowledge source unlike
the other top performing matching systems (AML,
LogMap and XMap). Table 1 illustrates the position-
ing of POMap (grey color) compared to the partici-
pated matching systems of OAEI 2016.
Conference. This track consists of matching seven
ontologies describing the conference organisation do-
main. Dealing with the conference dataset, we suc-
ceeded to achieve an F-Measure of 64%, which is an
average score compared to the other matching sys-
tems. Since this dataset includes the matching of
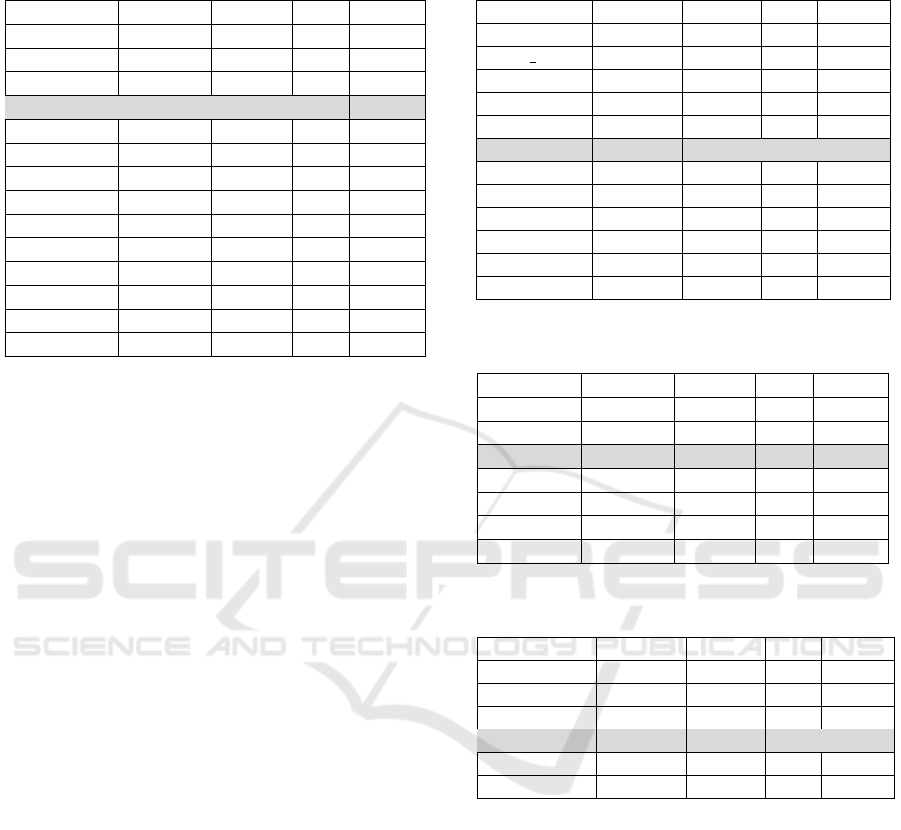
Table 1: POMap results in the anatomy track compared to
the OAEI 2016 systems.
System F-Measure Precision Recall Runtime
AML .943 .95 .936 47
CroMatcher .925 .949 .902 573
XMap .896 .929 .865 45
POMap .893 .931 .859 43
LogMapBio .892 .888 .896 11
FCAMAP .882 .932 .837 117
LogMap .88 .918 .846 24
LYAM .869 .863 .876 799
Lilly .83 .97 .794 272
LogMapLite .828 .962 .728 20
LPHOM .718 .709 .727 1601
ALIN .501 .996 .335 306
DkpAomLite .238 .99 .135 372
DkpAom .238 .99 .135 379
properties, we plan to add the data property and ob-
ject property matchers in our next version of POMap.
Large Biomedical Ontologies. The large Biomedical
Ontologies track aim to find the alignments between
three large ontologies: The Foundational Model of
Anatomy (FMA), SNOMED Clinical Terms and the
National Cancer Institute Thesaurus (NCI). Table 2
represents the obtained results for the FMA-NCI
small fragment. We succeed to achieve an F-measure
of 90%. Table 3 illustrates the results of POMap for
the FMA-SNOMED small fragment in which we ob-
tained an F-measure of 84% better than two matures
matching systems. We draws in the table 4 the results
of POMap in the SNOMED-NCI small fragment task
in which we achieved an F-measure of 73%. All the
resulted alignments by our system can be downloaded
at https://goo.gl/gRYtQj.
After performing the experimental process, we
can notice that some matching systems (AML,
LogMap, XMAP and Cromatcher) are performing
better than POMap. We argue that these systems are
outperforming POMap because of their implementa-
tion of a variety of matchers. However, POMap im-
plements only three matchers: one syntactic matcher
and two structural matchers. For instance, XMap em-
ploys different matchers such as a translation matcher
and a semantic matcher, which uses external back-
ground knowledge sources. However, POMap do not
perform any semantic matching process. Compared
to the runtime of other matching systems, we mention
that our element level matcher is the responsible of an
exponential growth of the execution time. However,
even with these drawbacks, we are able to accom-
plish acceptable results due to the efficiency of our
syntactic matcher resulted by a study of the different
similarity measures. Compared to the top perform-
Table 2: POMap results in the FMA-NCI small fragment
compared to the OAEI 2016 matching systems .
System F-Measure Precision Recall Runtime
XMAP .937 .977 .901 17
FCA MAP .935 .954 .917 236
AML .931 .963 .902 35
LogMap .924 .949 .901 10
LogMapBio .923 .935 .910 1712
POMap .900 .953 .852 241
LogMapLite .887 .967 .819 1
LYAM .796 .721 .889 1043
Lilly .657 .603 .721 699
ALIN .625 .995 .455 5811
DkpAom-Lite .615 .652 .577 1698
DkpAom .616 .652 .577 1547
Table 3: POMap results in the FMA-SNOMED small frag-
ment compared to the OAEI 2016 matching systems.
System F-Measure Precision Recall Runtime
XMAP .912 .989 .846 54
FCAMAP .865 .936 .803 1865
POMap .846 .928 .776 1223
AML .825 .953 .727 98
LogMapBio .801 .944 .696 1180
LOGMAP .799 .948 .690 60
LogMap .343 .968 .209 2
Table 4: POMap results in the SNOMED-NCI small frag-
ment compared to the OAEI 2016 matching systems .
System F-Measure Precision Recall Runtime
AML .797 .904 .713 537
LogMap .771 .922 .663 117
LogMapBio .770 .896 .675 3757
POMap .736 .813 .674 6920
XMap .697 .911 .564 .697
LOGMAPLite .693 .892 .567 693
ing systems of OAEI 2016, which are proposing sev-
eral matchers, POMap produces competitive results
using efficient matchers without relying on any exter-
nal knowledge resources. In order to have a better in-
sight of the efficiency of the structural matching step,
the following table 5 draws the results before and after
performing the structural matching process. The grey
color highlights the use of the structural matching.
5 CONCLUSION
In this paper, we described POMap a novel matching
system dedicated for the ontologies matching process.
POMap is employing syntactic techniques in order to
accomplish the element and structural level matching.

Table 5: POMap results before and after (grey color) using
the structural matching.
Track F-Measure Precision Recall
Anatomy .862 .942 .795
Anatomy .893 .931 .859
FMA-NCI (small) .897 .957 .844
FMA-NCI (small) .900 .953 .852
FMA-SNOMED (small) .836 .940 .752
FMA-SNOMED (small) .836 .928 .776
SNOMED-NCI (small) .736 .832 .651
SNOMED-NCI (small) .736 .813 .674
After performing a series of experiments, we provided
the top performing similarity measures and thresholds
that can be employed by an element level matcher
in the case of pre-processed ontologies. We have
proposed two structural matchers, which perform the
syntactic treatment over the siblings as well as the sub
classes. These two structural matchers explore the ini-
tial mappings resulted by the element level matcher in
order to improve the matching system results.
As a future work, we plan to adapt our matching
system in order to achieve a better run time over the
larger datasets of the OAEI campaign. While dealing
with other ontology matching fields rather than the
biomedical domain, other syntactic similarity mea-
sure can outperform the ones that we recommended.
Thus, we will concentrate on automating the process
of finding the best similarity measure and threshold
depending on the ontology matching context. There-
fore, we target the prediction of the optimal similar-
ity measure associated with its threshold by extracting
various features related to an ontology matching task.
REFERENCES
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nu-
cleic acids research, 32(suppl 1).
Cheatham, M. and Hitzler, P. (2013). String similarity met-
rics for ontology alignment. In International Semantic
Web Conference. Springer.
Cruz, I. F. and Sunna, W. (2008). Structural alignment
methods with applications to geospatial ontologies.
Transactions in GIS, 12(6).
Djeddi, W. E. and Khadir, M. T. (2014). A novel approach
using context-based measure for matching large scale
ontologies. In International Conference on Data
Warehousing and Knowledge Discovery. Springer.
Duan, S., Fokoue, A., and Srinivas, K. (2010). One size
does not fit all: Customizing ontology alignment using
user feedback. The Semantic Web–ISWC 2010.
Faria, D., Pesquita, C., Santos, E., Palmonari, M., Cruz,
I. F., and Couto, F. M. (2013). The agreementmak-
erlight ontology matching system. In OTM Confed-
erated International Conferences” On the Move to
Meaningful Internet Systems”. Springer.
Guli
´
c, M., Vrdoljak, B., and Banek, M. (2016). Cro-
matcher: An ontology matching system based on
automated weighted aggregation and iterative final
alignment. Web Semantics: Science, Services and
Agents on the World Wide Web, 41.
Jim
´
enez-Ruiz, E. and Grau, B. C. (2011). Logmap: Logic-
based and scalable ontology matching. In Interna-
tional Semantic Web Conference. Springer.
Megdiche, I., Teste, O., and Trojahn, C. (2016). An extensi-
ble linear approach for holistic ontology matching. In
International Semantic Web Conference. Springer.
Melnik, S., Garcia-Molina, H., and Rahm, E. (2002). Sim-
ilarity flooding: A versatile graph matching algorithm
and its application to schema matching. In Data Engi-
neering, 2002. Proceedings. 18th International Con-
ference on. IEEE.
Miller, G. A. (1995). Wordnet: a lexical database for en-
glish. Communications of the ACM, 38(11).
Monge, A. E., Elkan, C., et al. (1996). The field matching
problem: Algorithms and applications. In KDD.
Mungall, C. J., Torniai, C., Gkoutos, G. V., Lewis, S. E., and
Haendel, M. A. (2012). Uberon, an integrative multi-
species anatomy ontology. Genome biology, 13(1).
Nelson, S. J., Johnston, W. D., and Humphreys, B. L.
(2001). Relationships in medical subject headings
(mesh). In Relationships in the Organization of
Knowledge. Springer.
Ngo, D., Bellahsene, Z., and Todorov, K. (2013). Open-
ing the black box of ontology matching. In Extended
Semantic Web Conference. Springer.
Otero-Cerdeira, L., Rodr
´
ıguez-Mart
´
ınez, F. J., and G
´
omez-
Rodr
´
ıguez, A. (2015). Ontology matching: A litera-
ture review. Expert Systems with Applications, 42(2).
Porter, M. F. (2001). Snowball: A language for stemming
algorithms.
Schriml, L. M., Arze, C., Nadendla, S., Chang, Y.-W. W.,
Mazaitis, M., Felix, V., Feng, G., and Kibbe, W. A.
(2011). Disease ontology: a backbone for disease se-
mantic integration. Nucleic acids research, 40(D1).
Shvaiko, P. and Euzenat, J. (2013). Ontology matching:
state of the art and future challenges. IEEE Transac-
tions on knowledge and data engineering, 25(1).
Stoilos, G., Stamou, G., and Kollias, S. (2005). A string
metric for ontology alignment. The Semantic Web–
ISWC 2005.
Sun, Y., Ma, L., and Wang, S. (2015). A comparative eval-
uation of string similarity metrics for ontology align-
ment. Journal of Information &Computational Sci-
ence, 12(3).
