
Unsupervised Rib Delineation in Chest Radiographs by an
Integrative Approach
B. Buket Oğul
1
, Emre Sümer
2
and Hasan Oğul
2
1
Akgun Software Company, Ankara, Turkey
2
Department of Computer Engineering, Baskent University, Ankara, Turkey
Keywords: Computer Aided Diagnosis, X-ray Imaging, Medical Image Analysis.
Abstract: We address the problem of segmenting ribs in a chest radiography image as an intermediate step for
eliminating rib shadows for an effective Computer-Aided Diagnosis System (CAD). To this end, we
introduce a complete framework that takes an unprocessed x-ray image and reports the entire rib regions.
The system offers a novel strategy to fit a parabola curve to all rib seeds obtained through a log Gabor
filtering approach and extend the center curve by a problem-specific region growing technique to delineate
the entire rib, which does not necessarily follow a general parabolic model of rib cage. The visual
examinations of predicted rib delineations in a common dataset have demonstrated that the system can
achieve a reasonably good performance to be used in practice.
1 INTRODUCTION
Diagnostic image analysis is probably the biggest
contribution of computer and information sciences
to clinical practice in medicine. In the wake of
inferring abnormal structures from a medical image
without the intervention of a clinician, several
attempts have been done to develop intelligent
algorithms that can automatically analyze the raw
data from various types of modalities. X-ray
radiography is known as one of the cheapest and
least harmful of these imaging modalities. In spite of
the availability of a wide variety of more
complicated imaging techniques, it still remains as
ubiquitous in medical practice even for well-
equipped hospitals and radiology departments. On
the other hand, it is also evaluated as most difficult
modality to analyze, either manually or
computationally, due to the low resolution and high
noisy content of the input images.
Chest radiography is one of the most widely
used diagnostic tools in x-ray based imaging. A
common use is to detect potential lung nodules for
the decision of further examination using such as
computed tomography or pathology analysis. In
addition to major difficulties listed above, chest
radiography analysis suffers from normal anatomical
structures, such as bones and other overlapping
organs, which hide buried abnormalities in normal
tissue of lung. The nodules in the main tissue might
be invisible due to the shadows of quasi-parallel
bones in the ribcage. Therefore, the delineation and
suppression of ribs from the input image without any
information loss in the original lung tissue beneath is
an important computational challenge in computer-
aided diagnosis (CAD) systems for chest
radiographs. Whilst this study is motivated from our
ultimate goal of suppressing ribs for easier detection
of lung nodules, the automatic delineation of ribs has
several other benefits, such as providing a frame of
reference for locations of abnormalities or detecting
rib abnormalities, e.g. fractured or missing ribs. In
this study, we address the problem of segmenting the
anatomy of ribs in lung images obtained through the
x-ray radiography as an intermediate step of rib
shadow elimination in CAD systems. To this end,
we introduce a novel integrative approach based on
parabola curve fitting from rib seeds obtained
through a log Gabor filtering. The parabola is
extracted from a self-template. The system is
completely unsupervised, that is, it does not require
any training set of manually delineated ribs or any
knowledge-driven rules provided by an expert. The
visual examinations of predicted rib delineations in a
common dataset have demonstrated that the system
can achieve a reasonably good performance to be
used in practice.
The automated detection of ribs in chest
260
Ogul B., Sümer E. and Ogul H..
Unsupervised Rib Delineation in Chest Radiographs by an Integrative Approach.
DOI: 10.5220/0005361602600265
In Proceedings of the 10th International Conference on Computer Vision Theory and Applications (VISAPP-2015), pages 260-265
ISBN: 978-989-758-089-5
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

radiographs is indeed an old problem in medical
image analysis. In one of the earliest studies for rib
detection Toriwaki et al. (1980) located rib edges by
a template matching approach and fitted parabolas to
these borders. This idea later has been used in many
studies. De Souza (1983) used the derivatives of
vertical profiles to detect the pixels on the rib
boundaries and fitted parabola to these. Sarkar et al.
(1997) performed a set of gray-level calculation to
predict rib border candidates. Yue et al (1995) used
a modified Hough transform to fit parabolas to rib
edges and refined the boundaries by a snake
approach. Vogelsang et al. (1998) followed a similar
strategy for fitting parabolas to the edges by a
template matching approach and refined the ribs by
new estimates of parabolas for each rib. Karargyris
et al. (2011) smoothed the detected edges using a
Savitzk-Golay filter. Lee et al. (2012) used a
knowledge-based generalized Hough transform to
locate rib borders starting from a best template
detected by a simple Sobel operator. Horvath et al.
(2013) refined the final ribs by a dynamic-
programming-based active contour algorithm. In
general, all methods have three dimensions:
detecting the position of upper and/or lower rib
borders, fitting a curve to these borders, and refining
final rib shadows. It is also worth mentioning about
two supervised approaches that used a set of training
images with manually delineated rib borders
(Ginneken et al, 2000; Loog and Ginneken (2006))
and predicts the locations of ribs from a learned
model.
On the contrary to the existing methods that
employ a curve fitting from predicted upper or lower
rib edges, our system attempts to fit a template of a
parabola curve from a seed of rib start point which
can be more effectively predicted using a robust log
Gabor filtering approach. In this case, a curve
somewhere in the middle of the rib can be identified
instead of rib borders. Entire rib shadow is
delineated by a problem-specific region growing
technique, which is also introduced in this paper.
This growing stage allows the system to detect exact
rib regions which do not necessarily follow the fitted
parabola equation extracted from the best template
in the same image, which is considered as another
major contribution of this study.
The new system is visually examined in a well-
known chest radiography set (Shiraishi et al., 2000).
It is clearly demonstrated that the rib delineation can
be successfully achieved provided that at least one
template can be detected from original image. The
paper presents the steps of a complete framework
starting from several preprocessing stages including
lung segmentation, image enhancement and edge
detection and ending with final refinement stages for
rib delineation.
2 METHODS
2.1 Image Preprocessing
In image pre-processing step, we generate Local
Contrast Enhanced (LCE) and lung segmented
images to be used for template extraction and rib
detection steps. The details of these steps are not
given in here; a comprehensive description can be
found in (Ogul et al., 2015). Template extraction
step uses the product of lung segmented LCE and
original image as input.
2.2 Rib Segmentation
In this section we describe a novel hybrid approach
for rib segmentation. The existing systems which
model the ribs only using parabola or elliptic
equations are lack of the rib shape information
because it differs from person to person. We also
know that each person have a typical rib shape
which do not show significant changes. Therefore,
rather than trying to model the ribs regardless of the
shape changes, we propose to model the ribs using
existing rib templates which are extracted from the
overall lung field.
2.3 Rib Template Extraction
The rib detection task starts with the extraction of a
rib template from the content of query image itself.
This allows the framework to exhibit entirely in an
unsupervised manner. The template extraction
method is inspired from (Lee et al., 2012), where a
locale sampling scheme technique is first employed
to enhance the rib contrast, and then edge detection
operator is used to reveal candidate edges, and
finally a simple selection scheme is applied to fix the
template which appears in the middle of the lung
fields. The overall description of their methodology
can not detail how to convert edge detected image
into binary and how to select the template that
appears in the middle of the lung, where both
strategy in fact significantly affects the continuity of
the rib structure, and in turn the quality of the final
rib template extracted. In our study, we get the best
binary image results when we use Minimum Cross
Entropy (MCE) method for thresholding. However,
even the best thresholding technique results with
UnsupervisedRibDelineationinChestRadiographsbyanIntegrativeApproach
261

many small non rib candidates and does not provide
the rib continuity. Therefore, after MCE we apply
some morphological operators such as dilation to
connect disjointed ribs, opening to remove noise and
erosion to construct final rib structure (Fig 1).
Figure 1: Rib template extraction; first candidates (left),
after elimination stage (right).
To select the best candidates, we first flag 8
connected components which have highest pixel
count. Then, a thinning morphologic operator takes
place. At the final step we remove the templates by
their orientation, euler number and eccentricity
values. Remaining templates are further reduced
after detecting rib starts until we have one template
for each of the lung field, which is detailed in
following section.
2.4 Rib Delineation
The rib delineation through extracted self-templates
involves broadly 5 consecutive steps: (i) finding the
rib starts at outmost region of ribcage by a log Gabor
filtering, (2) selecting a single best template which is
most compatible with rib cage, (3) estimating rib
thickness, (4) parabola fitting to detect all ribs, and
(5) delineating the entire ribs by a specific region
growing technique.
2.4.1 Finding Outmost Rib Starts
Because the fact that the rib edges are easily affected
from high noise levels when pure pixel-based edge
detection methods used (Karargyris et al, 2011), we
choose to use Log Gabor wavelets in order to find
rib borders. Using just the 4
th
scale and the degrees
45° and 135° (Fischer et al., 2009) with Otsu
thresholding method gives us not all the rib
structures from vertical middle line to the outer
borders of the lung but the initial points with real
thickness values. In here, initial points refer to the
region in which the rib structure intersects with outer
border of the lung (Fig 2). Finally, by shifting the
lung mask we can see all the ribs positioned on
initial points (Fig 2) and we can eliminate the ones
which do not appear in initial points. For further
steps we also apply thinning morphologic operation
so that the ribs are formed as a single-pixel width.
Figure 2: Log Gabor filtering to detect rib seeds.
2.4.2 Selecting Best Template
In some cases, the method described in section 2.2.1
gives more than a one template for each of the lung
fields while sometimes we just get one template for
whole lung region. Thus we need to choose the best
template or to form a new one. When the first case
occurs, we reduce the templates by calculating their
distance to each of the thinned rib structures. The
closest template is chosen as the best template. In
cases such as a template for a lung field is found, but
calculated list for other field is empty, we form the
new template by taking the symmetry of existing
template.
Calculating the symmetry of a template is not an
easy task. It consists of mainly 3 steps: (i) First of
all, the best template must be formed as an elliptic
curve since it is comprised of scattered pixels (Fig 3,
dark blue lines on right lung). Using these non-
formed pixels, we find the coefficients for a
polynomial p(x) of degree n that is a best fit (in a
least-squares sense) for the data in y.
1
1
21
...)(
nn
nn
pxpxpxpxp
(ii) Secondly; its mirror symmetry is calculated (Fig
3, magenta template). (iii) And finally, its final
position is found with respect to the center of mass
of the region (Fig 3, yellow template). After we
compute the x and y coordinates of the vertex of the
parabola we found in step (i), we can easily calculate
the difference between this point and the center of
mass of the lung field in which this template lies on.
Since this distance should be same on the symmetric
lung field, by shifting the symmetric template as the
distance value gives us the final template on the left
lung in Fig 3 (yellow line).
2.4.3 Estimating Rib Thickness
Rib thickness (rt) value is used as a stopping
criterion while enlarging rib structure from its
VISAPP2015-InternationalConferenceonComputerVisionTheoryandApplications
262

center. The main point of calculating rt is to find the
longest vertical line that exists on rib structures.
Among all lines we calculate the mean of the values
except the ones which are too thin (
for JSRT < 10 px.)
or thick (>24 px.).
Figure 3: Best template selection step.
2.4.4 Parabola Fitting
Parabola fitting is very important task in which the
parameters of the parabola that fits the middle line of
each rib structure are calculated. This step is an
important basis for region growing and parabola
growing algorithms. In subsection 2.2.2.2 we
formulated both templates by calculating p
1
, p
2
, p
3
values. In order to fit the curve somewhere in the
middle of the rib, we need to optimize last parameter
of the parabola. While 1
st
and 2
nd
parameters are
shape related, 3
rd
parameter has an effect on the
position of the template on the rib by moving
template up and down. For each rib, keeping first 2
parameters same, we calculated list of p
3
values
using each pixel pair that appears in this structure
and their mean reports us the final position of the
template for its corresponding rib.
2.4.5 Rib Region Growing
Rib delineation is finalized with a two-stage region
growing approach. The first stage is the quasi-
parallel growing of initial parabola curve fitted to a
rib. We use the pixels in the middle of the rib as
initial seed points for region growing using 0.015 as
threshold value. The grown rib structure has broken
pixels on some points. We use these broken pixels
for generating our preliminary rib borders. The main
idea is repeating to shift the middle-line curve up
and down provided that shifted curve contains at
least 50% of initially grown pixels.
The first stage of region growing is resulted with a
ticker parabola. In some cases we can reach rib
borders on region growing part, however mostly the
result is thinner structure than actual rib. In the
second stage, we consider the rib consists of 4
horizontal parts and use each part individually in
order to form them into the final rib structure. For
each horizontal part, the growing process is repeated
separately on upper or lower part (not on both) based
on the mean intensity change until the exact rib with
predicted length is formed.
3 RESULTS
We compile our method on well-known JSRT
dataset having 154 chest x-ray images with known
nodules. In 150 of 154 images, at least one template
is identified and the best one is selected for self-
template matching. In 4 of 154 images, no template
is identified. This demonstrates the success of rib
template extraction step in general.
In 65% of images, all ribs are perfectly located
with a false discovery rate of 8.5%. This detection
rate is computed as 90.1% if only one rib is allowed
to be missed, which is usually the case since the
lowest left rib is often obscured by the heart shadow.
Here, we mean by rib detection the correction
identification of the position of the rib regardless of
that its borders are perfectly located. When each rib
is considered as a separate sample, the detection
recall (or sensitivity) is 97.1% with a precision of
91.5%. The delineation (locating the exact position
of all pixels) performance is not evaluated yet since
it requires the contribution of an expert knowledge,
which is currently in progress. Nevertheless, we
discern the ability of the system to delineate the ribs
by a few examples in Figure 4.
4 CONCLUSIONS
We introduce a complete framework that takes an
unprocessed lung image obtained thorough a chest
radiography and reports the pixels of entire rib
regions. The contribution of the study is two-fold.
First, a parabola equation extracted from a rib border
template is fitted into rib seeds which are defined as
the start points of the ribs closer to the outmost
(right or left) part of the ribcage. The previous
methods largely depend on the quantitative and
qualitative value of detected rib borders. Most often,
the method employed for edge detection may fail to
UnsupervisedRibDelineationinChestRadiographsbyanIntegrativeApproach
263

cover all ribs in the ribcage or report incorrectly
some other curves that resembles a rib boundary.
Since the present method does not require the
detection of any border but only its outmost starting
point, the detection rate can be substantially
increased by log Gabor filtering approach. Second
contribution is a new region growing technique that
is introduced to extend rib center to build the entire
rib until rib boundaries are reached. The technique
allows each individual rib to grow independently
from its initial parabola equation. In that sense, the
difference between the curvative structure of upper
and lower ribs can be elaborated. Based on the
visual examinations of a common dataset of lung
images, we can argue that the results are very
promising to continue with a suppression method
that follows the rib delineation.
The future work will involve: (1) a quantitative
evaluation the results after a manual delineation of
rib borders by an observer, (2) a qualitative
evaluation of the results by a committee of radiology
experts, (3) improving the algorithm that smoothes
the rib boundaries by a dynamic programming
approach, and finally (4) applying a data-driven
bone suppression technique to make the main tissue
more visible so that the abnormalities beneath the
ribs can be easily identified.
ACKNOWLEDGEMENTS
This study was supported by Turkey Ministry of
Science, Technology and Industry by the grant
number 379.STZ.2013-2, and Akgun Software
Company.
REFERENCES
De Souza P., Automatic rib detection in chest radiographs,
Comput. Vis., Graphics, Image Process., vol. 23,
pp.129 -161, 1983.
Fischer, S., Redondo, R. and Cristobal, G. “How to
construct Log-Gabor filters”, Open Access Digital
CSIC Document, 2009.
Horváth A., Orbán G.G., Horváth A., Horváth G., An X-
ray CAD system with ribcage suppression for
improved detection of lung lesions, Periodica
Polytechnica Electrical Engineering and Computer
Science, vol. 57, pp. 19-33, 2013.
Karargyris, A., Antani, S., Thoma, G., Segmenting
anatomy in chest x-rays for tuberculosis
screening, Engineering in Medicine and Biology
Society,EMBC, 2011 Annual International Conference
of the IEEE, On page(s): 7779 - 7782.
Lee J.S., Wangb J.W., Wuc H.H., Yuand M.Z., A
nonparametric-based rib suppression method for chest
radiographs Computers & Mathematics with
Applications, Volume 64, Issue 5, September 2012,
Pages 1390–1399.
Loog, M.; Ginneken, B. "Segmentation of the posterior
ribs in chest radiographs using iterated contextual
pixel classification", Medical Imaging, IEEE
Transactions on, On page(s): 602 - 611 Volume: 25,
Issue: 5, May 2006.
Ogul B.B., Kosucu P., Özcam A., Kanik S.D, Lung
Nodule Detection in X-Ray Images: A New Feature
Set, 6th European Conference of the International
Federation for Medical and Biological Engineering,
IFMBE Proceedings Volume 45, 2015, pp 150-155.
Sarkar S. and Chaudhuri S.,Detection of rib shadows in
digital chest radiographs, Proc. ICIAP (2), pp.356 -
363 1997.
Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T,
Kobayashi T, Komatsu K, Matsui M, Fujita H, Kodera
Y, and Doi K.: Development of a digital image
database for chest radiographs with and without a lung
nodule: Receiver operating characteristic analysis of
radiologists detection of pulmonary nodules. AJR 174;
71-74, 2000.
Toriwaki J., Hasegawa J, Fukumura T, Tagaki Y, (1980),
Computer analysis of chest photofluorograms and its
application to automated screening, Automedica 3, 63-
81.
van Ginneken B. and ter Haar Romeny V, Loew M. H.
and Sonka M., Automatic delineation of ribs in frontal
chest radiographs, Image Process., vol. 3979, pp.825
-836 2000.
Vogelsang F., Weiler F., Dahmen J., Kilbinger M.W.,
Wein B.B., Guenther R.W., Detection and
compensation of rib structures in chest radiographs for
diagnostic assistance, SPIE Proceedings Vol.3338:
Medical Imaging 1998: Image Processing.
Yue Z., Goshtasby A. and Ackerman L. V., Automatic
detection of rib borders in chest radiographs, IEEE
Trans. Med. Imag., vol. 14, no. 3, pp.525 -536 1995.
VISAPP2015-InternationalConferenceonComputerVisionTheoryandApplications
264
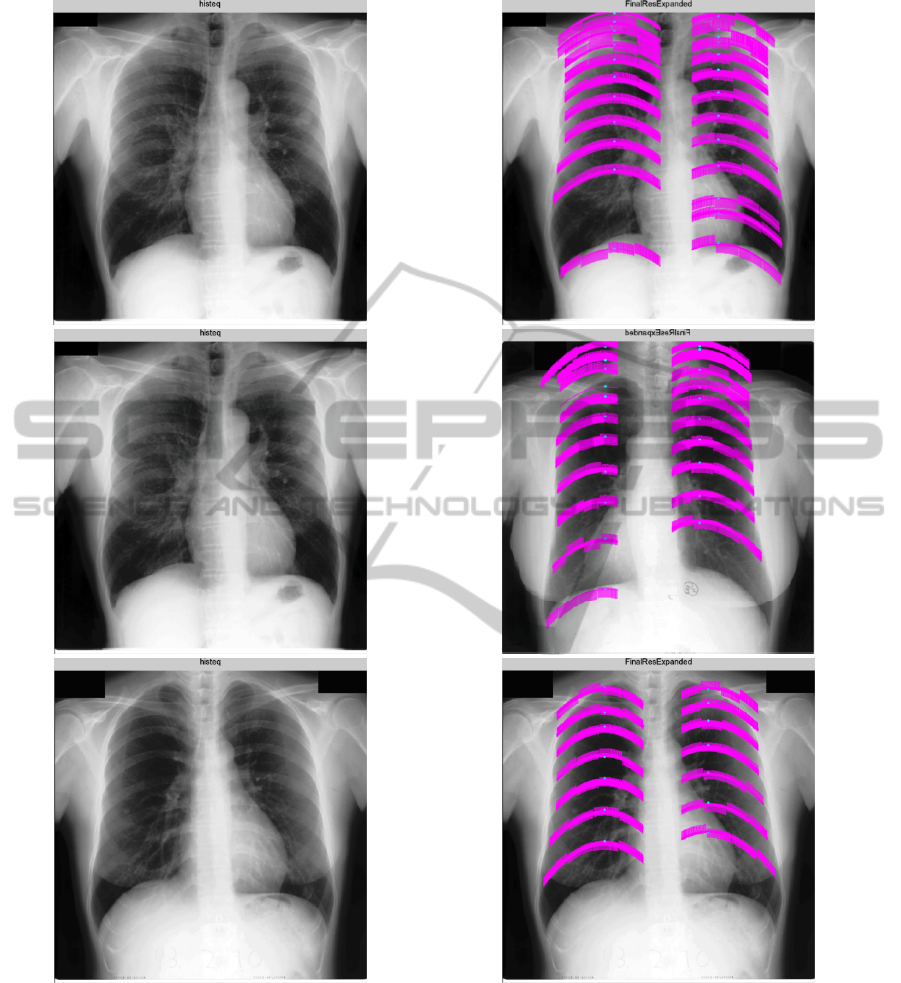
Figure 4: Example chest radiography images (left) and delineated ribs by our system (right).
UnsupervisedRibDelineationinChestRadiographsbyanIntegrativeApproach
265
