
Identifying Drug Repositioning Targets using Text Mining
Eduardo Barçante, Milene Jezuz, Felipe Duval, Ernesto Caffarena,
Oswaldo G. Cruz and Fabricio Silva
Fundação Oswaldo Cruz – Instituto Oswaldo Cruz, Av. Brasil 4365, Rio de Janeiro, Brazil
Keywords: Text Mining, Drug Repositioning, Clusters, PubMed Abstracts, Neglected Diseases, Ontology, Biomedical
Technology, Drug Industry, Information Extraction, Semi-Structured.
Abstract: The current scenario of computational biology relies on the know-how of many technological areas, with
focus on information, computing, and, particularly on the construction and use of existing Internet databases
such as MEDLINE, PubMed and PDB. In recent years, these databases provide an environment to access,
integrate and produce new knowledge by storing ever increasing volumes of genetic or protein data. The
transformation and management of these data in a different way than from the one that were originally
thought can be a challenge for research in biology. The problems appear by the lack of textual structure or
appropriate markup tags. The main goal of this work is to explore the PubMed database, the main source of
information about health sciences, from the National Library of Medicine. By means of this database of
digital textual documents, we aim to develop a method capable of identifying protein terms that will serve
as a substrate to laboratory practices for repositioning drugs. In this perspective, in this work we use text
mining to extract terms related to protein names in the field of neglected diseases.
1 INTRODUCTION
The improvement achieved in genomics research
can be seen as an excellent guidance to the new
social-economic dynamics that, among other politics
embraced by some countries, propose health as a
requirement for sustainable development. The
challenges proposed by the United Nations
declaration (2000) and the demand for new
information, which naturally arise from the
technological evolution, force the nations to
revaluate their health system research. The
orientation is that the results obtained in the
researches should be incorporated into health actions
and consequently reach sustainability of social
progress.
This way, it is necessary to acquire mechanisms
and strategies that can yield advances in the
biotechnological research and promote the essential
ways for them to be reused in the current knowledge
base. According to Markus, the necessary base,
composed by the raw material of innovation
processes, will bring improvements in health and in
all systems on which they depend on (Markus,
2001). However, a large number of challenges
follow these established set of goals, mainly in
biology, where biological databases present different
structures, representations and, standards (Schmitt,
2011). This conflicts with different cultures,
interdisciplinary models, technological limits and
urgent answers to the particular problems faced by
the population from each country.
The challenges rise in different contexts relative
to many productive sectors including the scientific
environment. An example of this is the rapid
increasing in the production of biological databases,
represented in the form of the scientific literature
that is widely publicized, articles, dissertations and
in genomics and protein databases too. Figure 1
shows the exponential growth of the scientific
production since the early nineties.
The motivation of this work is to explore this
kind of literature with information recovered from a
variety of databases, focused mainly on neglected
diseases (DNDi, 2010) such as malaria (WHO,
2013) and Chagas disease, which are substantial
issues that line up with the goals of the millennium
to be reached until 2015.
The neglected diseases can be regarded as a
group of diseases that can be found in tropical
regions (WHO 2010). However, up to the middle of
1970's, they were associated to places under extreme
348
Barçante E., Jezuz M., Duval F., Caffarena E., G. Cruz O. and Silva F..
Identifying Drug Repositioning Targets using Text Mining.
DOI: 10.5220/0005134903480353
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (KDIR-2014), pages 348-353
ISBN: 978-989-758-048-2
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)
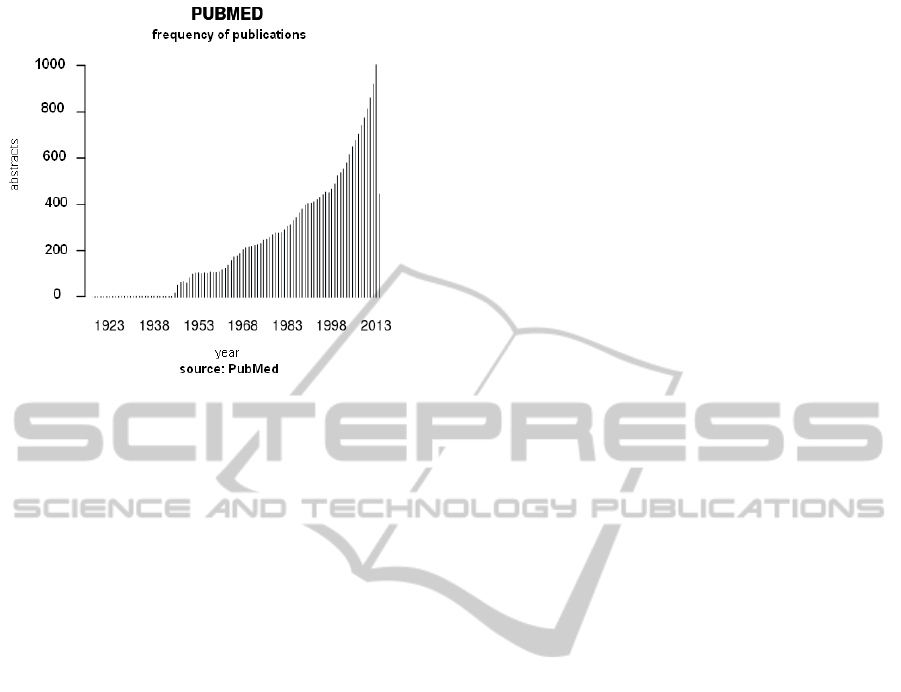
Figure 1: PubMed Frequency of the publications per year.
poverty. Today, they can be seen as a barrier in the
development of the countries located in the same
equatorial bands (INCT, 2014). They are called
“neglected” because the investments destined to
research, mainly by the pharmaceutics industry, do
not corroborate a sustainable cycle turned to
prevention and production of new drugs. (BRASIL,
2010).
In this article, we present a method to explore
digital textual documents deposited in PubMed
(PubMed, 2014). This methodology performs
queries in the digital documents selecting protein
names and relating them with protein databases with
the aim to form group of names of similar proteins,
in structure or function.
The identification of this set may provide inputs
for screening of molecules or may contribute with
data to drug repositioning.
Drug repositioning is defined on the basis of
descriptors in Health Sciences like deliberate and
methodical practice of finding new applications for
existing drugs (DECS, 2014).
James Black study (cited in Haupt, 2011)
anticipated that to achieve a new drug is necessary to
start from a known one: “The most fruitful basis for
the discovery of a new drug is to start with an old
drug.” As indicated by Wermuth (cited in Torres,
2014), to start from structural and therapeutically
diverse molecules, which have their bioavailability
and toxicity already tested, is the most beneficial
path to accomplish a new drug. Both authors defend
that the base of the method is the safety of a known
and approved drug providing considerable reduce of
time and cost in the testing phase. Many studies
performed by research groups at the Oswaldo Cruz
Foundation (FIOCRUZ) propose methods to
classify , organize and recover information in the
field of biotechnology, such as prioritization of
targets (Belloze, 2013); drug repositioning (Jardim,
2013); text mining (Jezuz, 2013), where they test
performance and similarity among biological and
scientific textual databases.
This paper is organized as following. In the
second and third sections, we will show a theoretical
framework that will guide the present work and
address the reuse, text mining and ontologies. The
proposed method will be explained in the fourth
section and the results in the fifth.
2 THE DATA REUSE AND TEXT
MINING
The reuse of data consists in the transformation and
the reorganization of data, in processes different
from the ones which they were originally designed
(Markus, 2001).
The problems appear due to the lack of a
standard record, textual structure or tags for
processing by computers, lack of appropriated
keywords to aid the process of searching and
recovering or by the large number of the possible
relations that must be built to individualize the
wanted information in more than one database, etc.
On the other hand, the reuse of technologies and
theories consolidated can be regarded as strong
points to the development of this work. It can be
quoted, the pioneer works of Swanson et al
(Swanson, 1986, 1987, 2006) that, by means of a
single counting of words could set a numerical
ranking, foreseeing significant mathematical and
quantitative relationships between datasets obtained
from the databases MEDLINE and Medical Subject
Heading (MeSH) (PubMed, 2014).
According to Feldman, extracting information is
one of the most important techniques used in text
mining, which is as a method of analyzing
unstructured texts or in natural language, with the
goal of recovering information and knowledge that
hardly would be achieved by humans in a single
reading (Feldman, 2002).
Therefore, text mining as a method of textual
analysis identifies and extracts information,
transforming texts in significant indexes intended to
prediction, clusters, etc. (Witten, 2004).
3 PROTEIN ONTOLOGY
Within the framework of neglected diseases, we will
IdentifyingDrugRepositioningTargetsusingTextMining
349

make a prospection of similar terms in the following
databases Protein Information Resource (PIR, 2014),
The Open Biological and Biomedical Ontologies
(OBO, 2014), Protein Data Bank (PDB, 2014) e
UniProt Consortium (UniProt, 2013). We will
employ methods for the formation of domains and
association of terms, such as controlled vocabularies
(Lancaster, 1986); descriptors and disjoint sets
(Swanson, 2006); Information Management
(Berners-Lee, 1990), among others.
We will get terms that set an ontological
representation (Campos et al, 2009) for proteins and
its explicit relationships to obtain different classes
that are displayed by agglomeration methods.
The assumption is based on the conjecture that
agglomerate information can establish a relevant
relationship related to the construction of the groups
that will be formed to assess the level of similarity
between the structures, functions and protein names.
4 RESEARCH METHODOLOGY
The project is divided into two phases: phase I sets
the programming framework, the terms that will be
used to search and recover abstracts in articles
deposited in PubMed database, the text mining. In
phase II, we will inspect for protein names in the
complete text articles previously selected, the search
for proteins with similar structure or function in
biological databases. And lastly to suggest inputs for
repositioning drugs.
4.1 Phase I
This work methodology will be developed using the
R programming language and environment (R
Foundation, 2002). It is a suitable free software
idealized to manipulate large amounts of data,
optimized to calculate and present results
graphically.
The data source will be PubMed. Currently, this
database contains more than 23 millions of
biomedical quoted literature of MEDLINE,
scientific journals and books online. Some citations
can include links towards the complete text content
of PubMed Central and sites of editors (PubMed,
2014).
The chosen terms will comprehend keywords,
correlated words to the topic or subjects related to
the query. Here, will use the following terms:
dengue, Chagas disease, malaria, leishmaniasis,
plasmodium and trypanosome.
The inputs will consist of abstracts collected in the
PubMed, written in their standard adopted format
(NLM, 2014) to semi-structured in R programming
language and environment (Feinerer, 2014).
The semi-structured data will be named as a
textual corpus or simply corpus.
The digital textual documents, in their raw
format, i.e. originated by metadata in XML format,
will require treatment for the formation of a textual
corpus (Feinerer, 2008), which needs to be modified
in a way to maintain in its content only the relevant
words to the proposed topic.
The preprocessing should be understood as an
initial phase in the text mining. First, the spurious
words that do not reflect the central theme are
removed. The objective is to extract a set of words
that represent all of a textual body that was
submitted to the natural processing of language. This
matrix term versus document follows the model of a
vector space (Salton, 1975) and has the purpose to
obtain a set of documents, their terms and their
respective frequencies. The third step is the analysis
and visualization of the data by means of clusters,
dendograms and word clouds, among other
techniques and functions.
Spurious data and stop words are terms that do
not translate the central theme of the text, such as
prepositions, articles, country name, slang, etc.
Consequently, they will be eliminated to obtain a
concise textual body, what will facilitate the
execution of the following procedures. It is
necessary to remove: a) words previously read in
the dictionary; b) country names, continents and
nationalities; c) prefix, suffix and verbs; d)
measurement units; e) terms identified during all the
processing that will not be in accordance with the
results obtained in the following phases. Therefore,
this group will form a new group of spurious terms
and will be verified and registered in the words
dictionary if needed.
In the present project, indexing and normalizing
the textual body will consist in disambiguate words
to reduce variability. The goal of this is to reduce to
a common term one set of words that have the same
sense or meaning.
The extracting terms will yield a set of words
after the processing of the textual corpus, in indexed
and normalized forms.
Finally, an analysis of the terms obtained in the
extracting process will be done to identify which
abstracts best represent the central topic that will be
representative of their corresponding full texts.
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
350
4.2 Phase II
Phase II will comprehend the search for protein
names in the complete article texts previously
selected along with the search for proteins with
similar structures or function in biological databases,
including proteins and ligands, to finally suggest
candidates for drug repositioning.
5 PRELIMINARY RESULTS
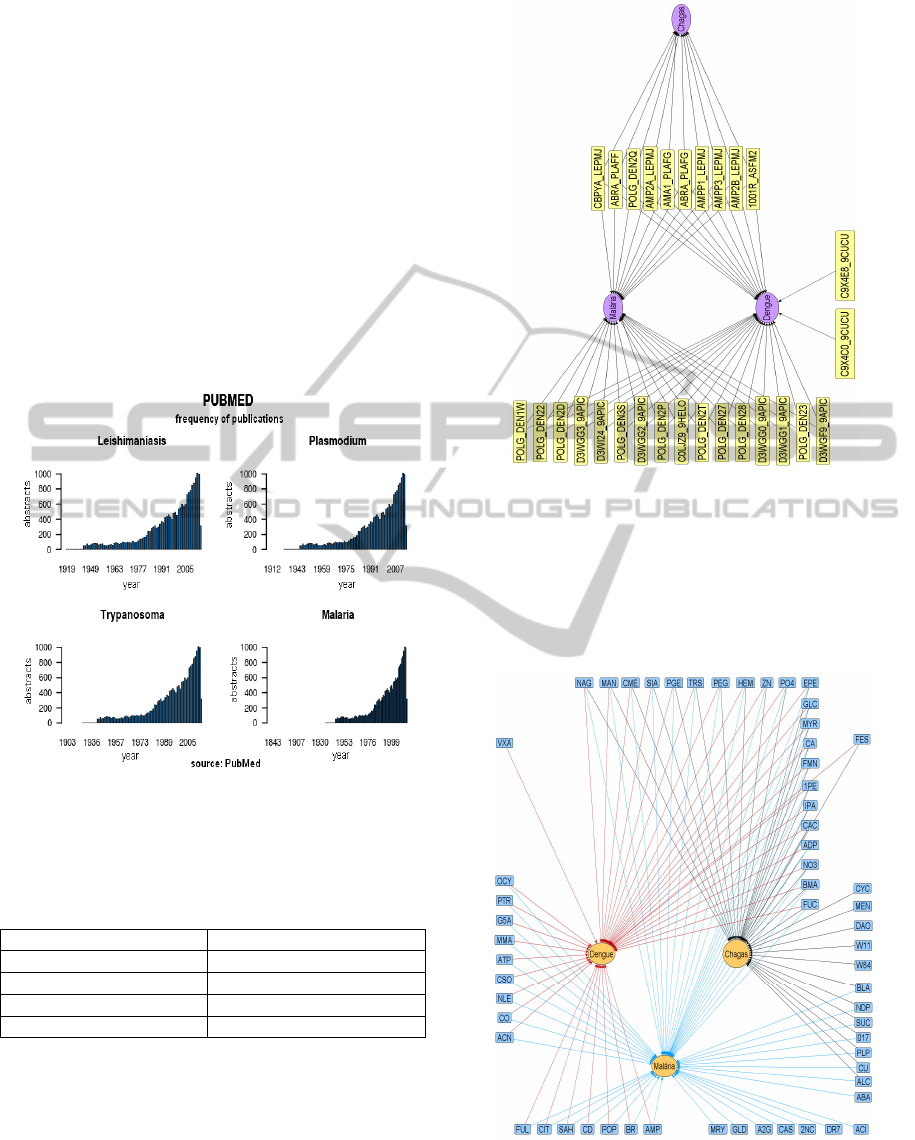
Figure 2 shows that the number of publications for
neglected diseases, Trypanosoma, malaria,
Plasmodium and leishmaniasis has been
continuously increasing the last years.
Figure 2: Neglected diseases terms in PubMed.
In table 1, there is a list of the files recovered
from the PubMed database and used in this work.
Table 1: Downloads summaries.
Terms Abstracts
Chagas disease 4.762
Dengue 7.524
Leishmaniasis 12.114
Malaria 30.298
Figure 3 shows the set of terms obtained from
abstracts retrieved from the Pubmed database, which
arose as results by means of the verification in the
protein databases PDB & UNIPROT. The outcomes
represent the crossing between proteins versus
disease names based on the frequency according to
which those terms appeared in the abstracts.
Figure 3: Proteins related to disease. Jezuz (2013).
With the proposed method it was possible to
collect the ligands related to the proteins names
found in UniProtKB. Figure 4 shows the set of
ligands and their relationship with extracted terms of
the articles listed regarding the neglected diseases
Dengue, Malaria and Chagas.
Figure 4: Ligands related to disease. Jezuz(2013).
From the ligands related to the proteins, it was
possible to obtain more information about the drugs
IdentifyingDrugRepositioningTargetsusingTextMining
351
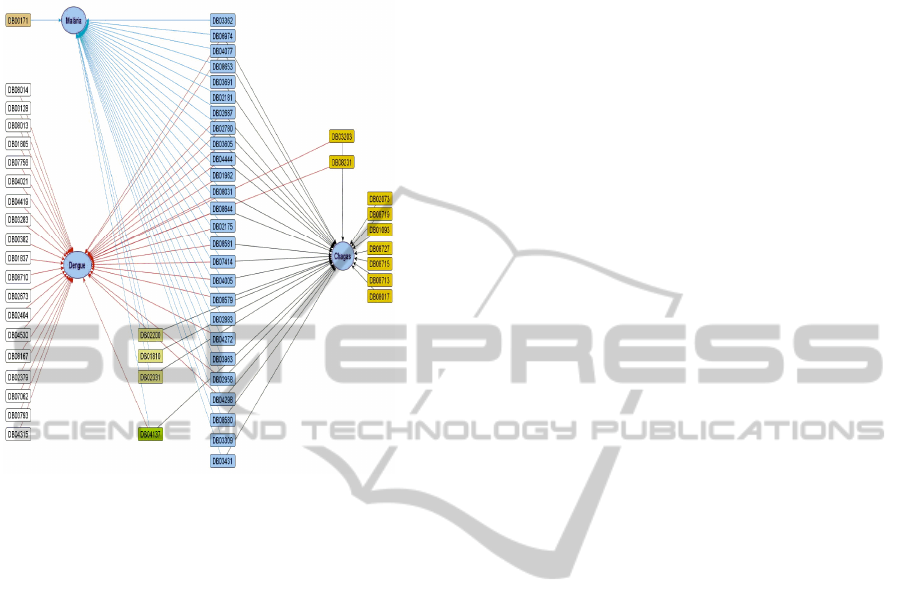
in the DrugBank database. The recovery of this
information enabled a link of drugs for neglected
diseases as can be seen in Figure 5.
Figure 5: Drugs related to disease. Jezuz (2013).
6 DISCUSSION
The proposed method is under construction, based
on theories and methods widely used and recognized
in the scientific scope. From the procedures related
with text mining, it was possible to reveal a way to
extract terms from the abstracts of scientific articles
and validate them as biological entities.
With graphs generation it was possible to
visualize relationships between data obtained from
pipeline execution using as inputs selections on the
matrix. An example of visualization is the
generation of graphs of terms related to diseases, and
graphs of proteins and similar structures. Figure 3
shows the graph containing relationships between
terms related to articles that have to do with
particular diseases (Chagas in this case). It also
shows those terms that were found in particular
articles on two or more diseases. Evidently, such
terms can just have been associated with different
diseases because they present common research
methodologies.
The relationships presented in Figure 4 show that
there are evidences that proteins related to ligands of
different articles could be part of studies which
assumption contemplates two or more diseases.
The "graph" also suggests that the ligands can
interact with diverse proteins present in organisms
related to different diseases.
Given the scope of this study, that information
can only be validated by experiments on the
laboratory.
Like the proteins and ligands previously
presented in Figure 5, there are several drug-related
to more than one disease. This relationship gives
the researcher the possibility to choose if the
application and study of drugs will be applied for
more than one disease.
So far, we have not exploited the methodology
using the full text articles. The main contribution of
this proposal is given by the fact of directly
contributing to the development and use of possible
arrangements of similar proteins in the structure of
function. It can also suggest the set for laboratory
practices as screening of molecules or its
applicability for drug repositioning.
ACKNOWLEDGEMENTS
This paper has been funded by project CAPES,
PAPES/VI.
REFERENCES
Belloze, K.T. 2013. Priorização de alvos para fármacos
no combate a Doenças Tropicais Negligenciadas
causadas por protozoários. FIOCRUZ/IOC. (In
Portuguese) [PhD thesis]
Berners-Lee T., 1990. Information Management: A
proposal. available from: http://www.w3.org/
History/1989/proposal.html [Accessed June 2014]
BRASIL. 2010. Ministry of Health. Neglected diseases:
the strategies of the Brazilian Ministry of Health. In
Journal of the Public Health. 2010;44(1):200-202.
Campos, M.L.A., Campos, M.L.M. 2009.
METHODOLOGICAL ASPECTS ON ONTOLOGY
REUSE: a study on the domain of trypanosomatids.
RECIIS – In R. Eletr. de Com. Inf. Inov. Saúde. Rio de
Janeiro, v.3, n.1, p.64-75, mar., 2009. online:
www.reciis.cict.fiocruz.br DOI: 10.3395/reciis.v3i1.
243en
DECS. 2014. Health Sciences Descriptors In Virtual
Health Library (VHL). available from http://decs.
bvsalud.org. [Accessed 08/08/2014]
DNDi. 2010. Drugs for neglected diseases initiative
available from: http://www.dndi.org.br/pt/doencas-
negligenciadas. [Accessed 08/08/2014]
Feinerer, I., 2008. A text mining framework in R and its
applications. [PhD thesis]. Vienna. Department of
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
352

Statistics and Mathematics, Vienna University of
Economics and Business Administration.
Feinerer, I., 2014. Text Mining Package available from:
http://cran.r-project.org
Feldman, R., Aumann, Y., Zilberstein, A., Ben-Yehuda,
Y., 2002. Mining biomedical literature using
information extraction. In Current Drug Discovery,
Volume2, Issue 10, pages 19-23,October 2002.
Gadelha, C.A.G., Quental C., Fialho B.C., 2003. Health
and innovation: a systemic approach in health
industries. In Reports in Public Health.
Haupt, V.J.; Schroeder, M. 2011. Old Friends in New
Guise:Repositioning of Known Drugs with Structural
Bioinformatics. In Brief. Bioinform. 12, 312−326.
doi:10.1093/bib/bbr011
INCT. 2014. National Institute for Science and
Technology on Innovation on Neglected Diseases
(INCT/IDN). Neglected Diseases. online: http://www.
cdts.fiocruz.br[Accessed 08/08/2014]
Jardim, R., 2013. Estudo de reposicionamento de fármacos
para Doenças Negligenciadas causadas por
protozoários através da integração de bases de dados
biológicas usando web semântica. FIOCRUZ/IOC. (In
Portuguese) [PhD thesis]
online: http://arca.icict.fiocruz.br/handle/icict/7027
Jezuz, M.P.G., 2013. Scientific text mining aiming at the
identification of bioactive compounds with
therapeutical potential against Chagas disease, dengue
and malaria. FIOCRUZ/IOC [PhD thesis]
Lancaster, F. W., 1986. Vocabulary control for
information retrieval. 2nd ed. Arlington, In VA:
Information Resources Press.
Markus L.M., 2001. Toward a theory of knowledge reuse:
Types of knowledge reuse situations and factors in
reuse success. In Journal of management information
systems, 2001;18(1):57-93.
NLM. 2014. MedLine® PubMed® XML Element
Descriptions and their Attributes available from:
http://www.nlm.nih.gov/bsd/licensee/elements_descrip
tions.html [Accessed June 2014]
OBO. 2014. The Open Bilogical and Biomedical
Ontologies.. online: www.obofoundry.org. [Accessed
08/08/2014]
PDB. 2014. Protein Data Bank. online: www.rcsb.org
PIR. 2014. Protein Information Resouce. online:
pir.georgetown.edu/pro/pro.shtml
PubMed, 2014. National Center for Biotechnology
Information online: http://www.ncbi.nlm.nih.gov
/pubmed. [Accessed 08/08/2014]
R Foundation, 2002. The R project for statistical
computing. online: http://www.r-project.org/
[Accessed 08/08/2014]
Salton, G., Wang, A., Yang, C.S., 1975. A vector space
model for information retrieval. Communications of
the ACM. 1975;18(11):613–620.
Schmitt, T., Messina, D.N., Schreiber, F., Sonnhammer
E.L.., 2011. Letter to the editor: SeqXML and
OrthoXML: standards for sequence and orthology
information. In Brief. Bioinform. 2011;12:485-488.
Swanson, D.R., 1986. Fish-oil, Raynaud’s syndrome and
undiscovered public knowledge. In Perspectives in
biology and medicine, 1986;30(1):7-18.
Swanson, D.R., 1987. Two medical literatures that are
logically but not bibliographically connected. In
Journal of the American society for information
science. 1987;38(4):228-333.
Swanson, D.R., Smalheiser, N.R., Torvik, V.I., 2006.
Ranking indirect connections in literature based
discovery. The role of Medical Subject Headings. In
Journal of the American Society for Information
Science and Technology, 2006;57(11):1427–39.
Torres, L.B. 2014. UFRJ. available from http://www.
portaldosfarmacos.ccs.ufrj.br/atualidades_profwermut
h.html [Accessed 08/08/2014]
UniProt Consortium, 2013. Update on activities at the
Universal Protein Resource (UniProt) in 2013. In
Nucleic Acids Res. 2013;41:D43-D47.
United Nations, 2000. Millennium development goals and
beyond 2015. online: http://www.un.org/millennium
goals/.[Accessed 08/08/2014]
WHO, 2013. Drugs for Neglected Diseases initiative
online: http://www.who.int
WHO, 2010. World Health Organization. First WHO
report on neglected tropical diseases: working to
overcome the global impact of neglected tropical
diseases. Geneva; 2010. online http://whqlibdoc.
who.int/publications [Accessed 08/08/2014]
Witten I.H, Don K.J, Dewsnip, M., Tablan V., 2004. Text
mining in a digital library. In Int J Digit Libr Journal.
2004;4:56-9.
IdentifyingDrugRepositioningTargetsusingTextMining
353
