
Chemical Master Equations
A Mathematical Scheme for the Multi-site Phosphorylation Case
∗
Alessandro Borri, Francesco Carravetta, Gabriella Mavelli and Pasquale Palumbo
Istituto di Analisi dei Sistemi ed Informatica “A. Ruberti”, Consiglio Nazionale delle Ricerche (IASI-CNR),
Viale Manzoni 30, 00185 Roma, Italy
Keywords:
Systems Biology, Chemical Master Equation, Markov Processes, Enzymatic Reactions.
Abstract:
The Chemical Master Equation (CME) provides a fruitful approach for the stochastic description of complex
biochemical processes, because it is able to cope with random fluctuations of the chemical agents and to fit
the experimental behavior in a more accurate way than deterministic concentration equations. In this work,
our attention is focused on modeling and simulation of multisite phosphorylation/dephosphorylation cycles,
by using the quasi-steady state approximation of enzymatic kinetics. The CME dynamics is written from the
coefficients of the deterministic reaction-rate equations and the stationary distribution is computed explicitly,
according to a recently developed realization scheme. Simulations are included to provide a comparison with
Monte Carlo methods in terms of computational complexity.
1 INTRODUCTION
Understanding complex cellular processes is a major
challenge facing present-day biology. Systems Bi-
ology is an approach that tackles the study of such
complex systems and is characterized by the fact that
molecular analysis - often at the genomic scale - is in-
tegrated with modeling, parameter identification, sim-
ulation analysis and control theory. Within this ap-
proach, biological processes are taken to be the re-
sults of complex, coordinated, dynamic, nonlinear in-
teractions of a large number of components, which
are affected by time and space constraints. The de-
velopment of efforts in the Systems Biology research
field will allow to elucidate relevant central features
of the more basic complex cellular functions (such
as cycle, growth, and so on) as well as to identify
their system-level properties, ultimately contributing
to shed more light on diseases arising as the impair-
ment metabolic functions such as cancer, diabetes,
etc. To this aim, a successful analysis of the com-
plex mechanisms arising in System Biology requires
the application of more and more efficient methods
for modeling such complex cellular systems.
As far as the networks of biochemical reactions,
there is an increasing interest in the development and
analysis of stochastic models, due to the preminent
∗
This work is partially supported by project IN-
TEROMICS.
role of fluctuations whenever a low number of
molecules is involved, (Mettetal and van Oudenaar-
den, 2007). Indeed, in a single cell environment, the
concentration of molecules can be low, so molecular
species can be found with a number of copies ranging
from hundreds to thousands. The small number of
molecules involved and the inherent stochasticity
of biochemical processes justify the necessity of
the stochastic approach to cope with intracellular
processes, (Van Kampen, 2007).
The biochemical complex machinery is largely
based on enzymatic reactions. Among the covalent
enzymatic-catalyzed modifications of substrates,
phosphorylation and dephosphorylation reactions
catalyzed by kinases and phosphatases, respectively,
provide a reversible post-translational substrate
modification that is central to cellular signalling
and regulation. In (Qu et al., 2003) it has been
proposed that multisite phosphorylation of tar-
get proteins by cyclin-dependent kinase proteins
is the likely source of nonlinear kinetic effects
in cell cycle control mechanisms. In particular,
dual phosphorylation/dephosphorylation cycles are
widely diffused within cellular systems and are
crucial for the control of complex responses such as
learning, memory, and cellular fate determination.
The majority of studies on biophysical analysis of
phosphorylation/dephosphorylation cycle have been
performed in an averaged, deterministic framework
681
Borri A., Carravetta F., Mavelli G. and Palumbo P..
Chemical Master Equations - A Mathematical Scheme for the Multi-site Phosphorylation Case.
DOI: 10.5220/0004632306810688
In Proceedings of the 3rd International Conference on Simulation and Modeling Methodologies, Technologies and Applications (BIOMED-2013), pages
681-688
ISBN: 978-989-8565-69-3
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

based on the Michaelis-Menten approach (Michaelis
and Menten, 1913). As stated above about generic
molecules, indeed in a single cell the substrate
and enzyme concentration can be low, being thus
necessary to study these cases within a stochastic
framework. Examples of biochemical reactions with
low number of substrates and involved enzymes
present at low concentrations are the phosphoryla-
tion/dephosphorylation cycle of AMPA-Receptors
(AMPARs) in a single synaptic spine and have been
treated in (Castellani et al., 2001; Whitlock et al.,
2006). In (Castellani et al., 2001) it has been pro-
posed a synaptic receptors model (AMPAR model)
which, at a molecular-microscopic level, is a candi-
date for memory storage and switching behavior, and
a kinetic scheme of the enzymatic reactions describ-
ing the AMPAR double phosphorylation channel has
been given. It has been stressed that the AMPAR
modification, a 2-step phospho/dephosphorylation
cycle, guaranties the synaptic plasticity and exhibits
bistability for a wide range of parameters, consistent
with values derived from biological literature.
In this paper we investigate the Chemical
Master Equation (CME) approach applied to
model in a stochastic framework the phosphoryla-
tion/dephosphorylation cycle. Indeed, the CMEs
provides an appropriate description of complex cellu-
lar processes, (Van Kampen, 2007; Gillespie, 1977).
It is a powerful method even with significant sim-
plifications, such as spatial homogeneity of volumes
where the chemical reactions under investigation are
taking place. The CME is a promising approach for
Systems Biology modelling for a variety of reasons,
such as its capability of coping with fluctuations and
chemical fluxes and of well fitting experimental data
in the today widespread single cell experiments, and
its capability to capture and explain the deviation
from Gaussianity observed in various gene expression
experiments (such as stress or metabolic response,
growth of the nuclear protein amount observed in
senescent cells, and so on). It results more infor-
mative of the real behavior of the system than the
deterministic approach, since the results obtained in
a deterministic framework cannot describe diffusion
effects due to fluctuations and chemical currents
capable to drive switching from one equilibrium to
the other. Such mechanism has a relevant role also
in neuronal plasticity phenomena, (Castellani et al.,
2009). In (Bazzani et al., 2012) the stationary dis-
tribution provided by the two-dimensional chemical
master equation for a well-known model of a two
step phosphorylation/dephosphorylation cycle has
been studied, by using the quasi-steady state approx-
imation of enzymatic kinetics. Authors pointed out
the possibility for the molecule distribution shape to
be controlled (in particular changed from a unimodal
distribution to a bimodal distribution) by chemical
fluxes occurring in the biochemical system under
investigation. This phenomenon corresponds to the
typical mechanism that is adopted by such system
to obtain a plastic behavior, for example when any
kind of exogenous chemical messenger is present, by
changing the properties of the biochemical reaction
occurring inside the cell.
The paper is organized as follows: in Section 2 we
review some structural properties of the Master Equa-
tion and address the efficient computation of its equi-
librium solution. In Section 3, we illustrate the case
study of multiple phosphorylation for an arbitrary
number of sites. In Section 4 we propose some sim-
ulation results concerning the case of a triple phos-
phorylation framework. Section 5 offers concluding
remarks.
2 THE CHEMICAL MASTER
EQUATION
Consider M (bio)chemical species Y
1
,...,Y
M
involved
in q chemical reactions (Ullah and Wolkenhauer,
2011) described by the stoichiometric coefficients β
ij
,
for any species j = 1,...,M and for any reaction i =
1,...,q. The Chemical Master Equation (CME) de-
scribes the time evolution of the probability of being
in one of the N ≤ +∞ possible configurations (states)
at any time (Van Kampen, 2007). More generally, the
CME is the dynamic equation of a continuous-time
discrete-state stochastic Markov process. If N is fi-
nite, the equations for the joint probabilities can be
collected in the form of an autonomous positive lin-
ear system (Farina and Rinaldi, 2000):
˙
P = GP , P ∈ R
N
, (1)
where G is called infinitesimal generator of the
Markov process.
We define as n(t) = (n
1
(t),... , n
M
(t)) the state of
the system at time t, with i-th component n
i
(t) ∈ N
0
denoting the number of copies of species Y
i
at time
t. The generic element G
ij
of G is the propensity
g
α
1
,...,α
M
n
1
,...,n
M
of reaching the state v
j
= (n
1
+ α
1
,...,n
M
+
α
M
) from the state v
i
= (n
1
,...,n
M
).
Depending on whether or not the system of re-
actions is closed (the total mass is conserved) and
according to the number of distinct elements form-
ing the M species, the values of some components of
the vector state n(t) can be inferred from the others,
which makes the state representation redundant. We
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
682

will henceforth assume that the M species are inde-
pendent, i.e. the state representation is minimal (non-
redundant). The interested reader is referred to (Borri
et al., 2013) for a deeper discussion on these and on
the following concepts.
Note that different ways of aggregation of the
probabilities in the vector P may produce different
properties of the linear system in (1). In particular,
one can consider the following recursive partitioning:
P
n
1
,...,n
M−1
.
=
p
n
1
,...,n
M−1
,0
p
n
1
,...,n
M−1
,1
.
.
.
p
n
1
,...,n
M−1
,N
M
∈ R
N
M
+1
, (2)
with 0 ≤ n
i
≤ N
i
, with N
i
being the maximum number
of allowed copies of species Y
i
, and p
n
1
,n
2
,...,n
M
being
the joint probability of having n
i
copies of species Y
i
,
for all i = 1,...,M. Then, the following vectors of
probabilities can be recursively defined
P
n
1
,...,n
i
.
=
P
n
1
,...,n
i
,0
P
n
1
,...,n
i
,1
.
.
.
P
n
1
,...,n
i
,N
i+1
∈ R
(N
i+1
+1)×···×(N
M
+1)
,
(3)
for 1≤ i≤ M−2. This leads to the definition of vector
P , entailing all the joint probabilities involved by the
CME:
P
.
=
P
0
P
1
.
.
.
P
N
1
∈ R
(N
1
+1)×···×(N
M
+1)
. (4)
The way of collecting the probabilities in P as in
(2)–(4) induces a recursive block partitioning on ma-
trix G, according to (1). In case the stoichiometric
coefficients satisfy the unitary step property:
β
ij
∈ {−1,0, 1},i ∈ {1,...,q}, j ∈ {1,...,M}, (5)
an interesting recursivestructure of the CME dynamic
equations can be pointed out:
Proposition 1. Consider a continuous-time Markov
process describing a set of q chemical reactions in-
volving M independent (bio)chemical species and de-
scribed by a set of stoichiometric coefficients {β
ij
}
satisfying (5). Then the infinitesimal generator G
has the following block-tridiagonal structure (see e.g.
(Carravetta, 2011)):
G=Φ
N
1
{G
1
n
1
},{G
0
n
1
},{G
−1
n
1
};n
1
=
G
0
0
G
−1
1
/
0 ··· ···
/
0
G
1
0
G
0
1
G
−1
2
/
0 ···
/
0
/
0
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
/
0
.
.
.
.
.
.
.
.
.
G
1
N
1
−2
G
0
N
1
−1
G
−1
N
1
/
0 ··· · · ·
/
0 G
1
N
1
−1
G
0
N
1
,
(6)
where the
/
0 entries are zero matrices of proper dimen-
sions. Furthermore, all the non-zero blocks of G are
block-tridiagonal with the same structure of G.
The operator Φ
ν
({A
i
},{B
i
},{C
i
};i) used in (6) is
called block-tridiagonal matrix builder and is defined
as follows
Φ
ν
({A
i
},{B
i
},{C
i
};i) :=
B
0
C
1
/
0 ···
/
0
A
0
B
1
C
2
/
0
/
0
/
0
.
.
.
.
.
.
.
.
.
/
0
.
.
.
.
.
.
A
ν−2
B
ν−1
C
ν
/
0 ···
/
0 A
ν−1
B
ν
,
(7)
where {A
i
}, {B
i
}, {C
i
} are sequences of suitably
dimensioned square matrices, and the
/
0 entries are
zero matrices of proper dimensions. A network of
chemical reactions satisfying (5) allows simultane-
ous changes of unitary amount in the state variables
and is later referred as generalized one-step process
or unitary process. It is possible to show that (5) is
the mildest condition preserving the recursive block-
tridiagonal structure of G.
The main consequenceof Proposition 1 is that G is
sparse, which reveals to be useful to carry out classi-
cal tasks as the computation of the stationary solution
of the CME, satisfying
GP = 0. (8)
The following result, based on an interpretation of
the biochemical network in terms of Algebraic Graph
Theory (Bullo et al., 2009), characterizes the prop-
erty of uniqueness of the CME stationary distribution,
which makes the study of the equilibrium problem (8)
independent of the initial state.
Proposition 2. The stationary distribution P
ss
of a
continuous-time Markov process is unique if and only
if the digraph associated with the Markov process has
a globally reachable vertex. Under this assumption, 0
is a simple eigenvalue of G with right eigenvector u
0
and the stationary distribution is given by P
ss
=
u
0
1
T
u
0
.
ChemicalMasterEquations-AMathematicalSchemefortheMulti-sitePhosphorylationCase
683

As an alternative approach to the computation of
P
ss
in terms of the right eigenvector u
0
of G, Propo-
sitions 1 and 2 allow an efficient computation of the
solution of (8) by means of Gaussian elimination:
Proposition 3. The algorithm of Gaussian elimi-
nation to solve the stationary equation GP = 0 in
unitary-step processes is performed in time O (N
2
),
where N is the number of rows of G.
Note that the complexity of the Gaussian Elimina-
tion for dense matrices is O(N
3
).
3 MULTI-SITE
PHOSPHORYLATION
This section is devoted to formalizing, in the CME
mathematical setting, the multi-site phosphorylation
reaction framework.
3.1 Writing the Reaction-rate
Equations
Consider the case of only M = 2 phosphorylationsites
(see e.g. (Bazzani et al., 2012)), depicted in Fig. 1,
involvingM+1= 3 species and 2 enzymes (kinase E
1
and phosphatase E
2
). Such a framework is formalized
by the following set of q = 2M = 4 reactions:
A
(0)
−→ A
(1)
,
A
(1)
−→ A
(2)
,
A
(1)
−→ A
(0)
,
A
(2)
−→ A
(1)
,
where the species A
(0)
, A
(1)
, A
(2)
are the
non-phosphorylated, phosphorylated and double-
phosphorylated substrate, respectively. If one con-
siders the double-site phosphorylation as a closed
system (the total substrate concentration is constant
[A
(0)
]+ [A
(1)
]+ [A
(2)
] = A
tot
), the number of indepen-
dent species is M = 2.
We now address the general M-site phosphoryla-
tion case, illustrated in Fig. 2. We can consider the
case of closed system, so that the species A
(i)
may be
intended to be the i-times phosphorylated substrate,
for i = 0,...,M, and the total substrate concentration
is constant
∑
M
i=0
[A
(i)
] = A
tot
. The odd-indexed reac-
tions are catalyzed by the kinase E
1
:
[A
(i−1)
] + [E
1
]
K
2i−1
ON
↼−−−
−−−⇁
K
2i−1
OFF
[A
(i−1)
E
1
]
v
2i−1
−−−→ [A
(i)
] + [E
1
]
(9)
with i = 1, ..., M, while the even-indexed reactions
are activated by the phosphatase E
2
:
[A
(i)
] + [E
2
]
K
2i
ON
↼−−−
−−−⇁
K
2i
OFF
[A
(i)
E
2
]
v
2i
−→ [A
(i−1)
] + [E
2
] (10)
with i = 1,...,M, for a total number of reactions
equal to q = 2M.
The left-hand M pairs of reactions in (9)–(10) are
considered at the equilibrium (quasi-steady-state hy-
pothesis), thus the corresponding 2M deterministic
Michaelis-Menten (MM) equations rewrite:
d[A
(i−1)
E
1
]
dt
=K
2i−1
ON
[A
(i−1)
][E
1
]−K
2i−1
OFF
[A
(i−1)
E
1
]=0
d[A
(i)
E
2
]
dt
=K
2i
ON
[A
(i)
][E
2
]−K
2i
OFF
[A
(i)
E
2
]=0
for i = 1,...,M. Hence, by setting the dissociation
coefficients k
j
:= K
j
OFF
/K
j
ON
, j = 1,...,2M, one gets
the equilibrium conditions:
[A
(i−1)
E
1
]
eq
=
[A
(i−1)
][E
1
]
k
2i−1
[A
(i)
E
2
]
eq
=
[A
(i)
][E
2
]
k
2i
for i = 1,...,M. We now focus on the right-hand
reactions in (9)–(10), where we replace the terms
[A
j
E
k
] with their equilibrium values [A
j
E
k
]
eq
, for j =
1,...,2M and k = 1,2. One gets:
[A
(i−1)
][E
1
]
k
2i−1
v
2i−1
−−−→ [A
(i)
] + [E
1
], (11)
[A
(i)
][E
2
]
k
2i
v
2i
−→ [A
(i−1)
] + [E
2
]. (12)
Note that the generic species A
(i)
, for i = 1,...,M, ap-
pears in 4 distinct reactions:
[A
(i−1)
][E
1
]
k
2i−1
v
2i−1
−−−→ [A
(i)
] + [E
1
],
[A
(i)
][E
2
]
k
2i
v
2i
−→ [A
(i−1)
] + [E
2
],
[A
(i)
][E
1
]
k
2i+1
v
2i+1
−−−→ [A
(i+1)
] + [E
1
],
[A
(i+1)
][E
2
]
k
2(i+1)
v
2(i+1)
−−−→ [A
(i)
] + [E
2
],
which lead to the following MM equation, for any i:
d[A
(i)
]
dt
=
v
2i−1
k
2i−1
[E
1
][A
(i−1)
] −
v
2i
k
2i
[E
2
][A
(i)
]
−
v
2i+1
k
2i+1
[E
1
][A
(i)
] +
v
2(i+1)
k
2(i+1)
[E
2
][A
(i+1)
]. (13)
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
684

E
1
E
1
E
2
E
2
1
3
2
4
A
(0)
A
(1)
A
(2)
Figure 1: Double-site Phosphorylation.
A
E
1
E
1
E
2
E
2
1
3
2 4
E
1
2i − 1
2i
E
1
E
2
2i + 1
2(i + 1)
E
1
E
1
E
2
E
1
E
1
(0)
A
(1)
A
(i)
A
(M −1)
A
(M )
E
2
E
2
2M − 1
2M
2M − 3
2M − 2
Figure 2: Multi-site Phosphorylation.
Note that i = 0, i = M are special cases because some
terms are neglected:
d[A
(0)
]
dt
= −
v
1
k
1
[E
1
][A
(0)
] +
v
2
k
2
[E
2
][A
(1)
], (14)
d[A
(M)
]
dt
=
v
2M−1
k
2M−1
[E
1
][A
(M−1)
] −
v
2M
k
2M
[E
2
][A
(M)
].
(15)
The following mass-balance constraints on the total
substrate concentration and on the enzymes:
M
∑
i=0
[A
(i)
] = A
tot
,
[E
1
] +
M
∑
i=1
[A
(i−1)
]
k
2i−1
[E
1
] = E
1,tot
,
[E
2
] +
M
∑
i=1
[A
(i)
]
k
2i
[E
2
] = E
2,tot
,
(16)
lead to the a minimal representation of the process by
means of M independent species. For instance, we
choose x
i
= [A
(i)
] for i = 1,...,M and, from (13), we
are able to describe the system in terms of M coupled
differential equations, called reaction-rate equations:
˙x
1
=
v
1
k
1
E
1
(x)x
0
−
v
2
k
2
E
2
(x) +
v
3
k
3
E
1
(x)
x
1
+
v
4
k
4
E
2
(x)x
2
.
.
.
˙x
i
=
v
2i−1
k
2i−1
E
1
(x)x
i−1
−
v
2i
k
2i
E
2
(x) +
v
2i+1
k
2i+1
E
1
(x)
x
i
+
v
2(i+1)
k
2(i+1)
E
2
(x)x
i+1
.
.
.
˙x
M
=
v
2M−1
k
2M−1
E
1
(x)x
M− 1
−
v
2M
k
2M
E
2
(x)x
M
(17)
where x
0
:= [A
(0)
] and E
1
(x), E
2
(x) are given by:
x
0
= 1 −
M
∑
i=1
x
i
,
E
1
(x) =
E
1,tot
1+
∑
M
i=1
x
i−1
k
2i−1
,
E
2
(x) =
E
2,tot
1+
∑
M
i=1
x
i
k
2i
.
(18)
3.2 From the Reaction-rate Equations
to the Master Equation
In (17)–(18), we built a minimal deterministic repre-
sentation of the M-site phosphorylation by means of
a system of M ordinary differential equations (ODEs)
with respect to the M variables x
1
,...,x
M
, where x
i
represents the normalized concentration of species
Y
i
, i.e. of the i-times phosphorylated substrate. It
is deeply discussed in the related literature how the
reaction-rate approach fails to capture the inherent
randomness of biochemical phenomena, especially in
the presence of a low number of molecules (Van Kam-
pen, 2007). In the following, we build a stochas-
tic representation of the chemical process in terms of
Master Equations.
A common approach to infer the CMEs from the
ODEs consists in turning the real-valued concentra-
tion variables x
i
into the integer values n
i
, represent-
ing the copies of species Y
i
, according to the mass-
balance constraint:
n
0
= n
TOT
−
M
∑
i=1
n
i
.
As a first step, we need to write the stoichiometric
coefficients for each reaction.
Odd-indexed Reactions. The generic reaction
2i − 1, for i = 1,...,M, “generates” a molecule of
species Y
i
and “annihilates” a molecule of species
Y
i−1
, from (11). Hence the vector of stoichiometric
ChemicalMasterEquations-AMathematicalSchemefortheMulti-sitePhosphorylationCase
685

coefficients
¯
β
2i−1
= (β
2i−1,1
,...,β
2i−1,M
) for these re-
actions is given by:
β
2i−1, j
=
1 j = i,
−1 j = i− 1,
0 otherwise.
(19)
Even-indexed Reactions. The generic reaction 2i,
for i = 1,...,M, reverses the reaction 2i− 1, namely it
generates a molecule of species Y
i−1
and annihilates
one of species Y
i
, from (12). Hence the vector of stoi-
chiometric coefficients
¯
β
2i
= (β
2i,1
,...,β
2i,M
) for these
reactions is given by:
β
2i, j
=
−1 j = i,
1 j = i− 1,
0 otherwise.
(20)
Writing the Matrix G. We are now ready to
write the CME by computing the generic element g
α
n
(propensity), with the shortcuts n = (n
1
,...,n
M
) and
α = (α
1
,...,α
M
). By comparison with the equation of
species i in (17):
˙x
i
=
v
2i−1
k
2i−1
E
1
(x)x
i−1
−
v
2i
k
2i
E
2
(x) +
v
2i+1
k
2i+1
E
1
(x)
x
i
+
v
2(i+1)
k
2(i+1)
E
2
(x)x
i+1
(21)
and taking into account the definition of the vectors of
stoichiometric coefficients in (19)–(20), one can de-
fine:
g
α
n
:=
v
2i−1
k
2i−1
E
1
(n)n
i−1
α =
¯
β
2i−1
,i = 1,...,M,
v
2(i+1)
k
2(i+1)
E
2
(n)n
i+1
α =
¯
β
2(i+1)
,i = 1,...,M,
−
M
∑
i=1
v
2i
k
2i
E
2
(n)+
v
2i+1
k
2i+1
E
1
(n)
n
i
α = 0,
0 otherwise,
(22)
where 0 = (0,...,0). Intuitively, the definition in (22)
is obtained from (21) by regarding the positive ad-
dends as generation terms (i.e. creating a molecule)
and the negativeones as recombination terms (i.e. an-
nihilating a molecule) of the associated biochemical
reactions. The dynamic matrix G of the CME can be
written as in (6), where the scalar blocks are defined
from the propensities as:
G
α
1
,...,α
M
n
1
,...,n
M
=
g
α
1
,...,α
M
n
1
,...,n
M
if (α
1
,...,α
M
) 6= 0,
−
∑
¯
α
1
,...,
¯
α
M
g
¯
α
1
,...,
¯
α
M
n
1
,...,n
M
otherwise,
(23)
and the general blocks are recursively defined in
terms of the matrix builder in (7) as:
G
α
1
,...,α
i
n
1
,...,n
i
=Φ
N
i+1
{G
α
1
,...,α
i
,1
n
1
,...,n
i
,n
i+1
},
{G
α
1
,...,α
i
,0
n
1
,...,n
i
,n
i+1
},{G
α
1
,...,α
i
,−1
n
1
,...,n
i
,n
i+1
};n
i+1
(24)
for 1≤ i ≤ M−1, 0 ≤ n
i
≤ N
TOT
, 0 ≤ |α
i
| ≤ N
TOT
.
It can be shown (see e.g. (Van Kampen, 2007))
that the deterministic reaction-rate approach is a first-
order approximation of the dynamics of the mean
value of the CME.
4 SIMULATION RESULTS
In the following, we restrict our attention to the case
M = 3 (triple phosphorylation) and we compare the
performance between computing the stationary distri-
bution explicitly, based on the results in Section 2, and
obtaining it by means of statistical methods, in par-
ticular the Gillespie Stochastic Simulation Algorithm
(SSA) (Gillespie, 1977).
For M = 3, we have q = 6 reactions. We assume
n
TOT
= 40 and E
1,TOT
= E
2,TOT
= 1. The dimen-
sion of G is N = 41
3
> 64, 000, but an inspection
of the associated states reveals that a large subset of
those are not reachable (in the sense of Munsky et al.
(Munsky and Khammash, 2008)) because they vio-
late the mass-balance constraints, hence they have a
zero steady-state probability. If one erases a priori
the components referred to those states from the mas-
ter equation, a reduced matrix
˜
G is obtained, whose
kernel is 1-dimensional. This ensures the uniqueness
of the stationary distribution
˜
P
ss
, defined over the set
of reachable states of the Markov Process. In this par-
ticular case, just
˜
N = 12, 341 configurations are ad-
missible states.
The values of the chosen parameters are: k
1
=
k
6
= 1, k
2
= k
3
= 1.8, k
4
= k
5
= 2.2, v
1
= v
2
= 1,
v
3
= v
4
= 1.1, v
5
= v
6
= 1.2. We apply the formaliza-
tion in Section 3 and perform numerical simulations
in the Matlab suite on an Apple MacBook Pro laptop
with 2.5 GHz Intel Core i5 CPU and 16 GB RAM.
We started with the stochastic simulation. We
considered as initial state n(0) = (0, 0,0), meaning
that n
0
= n
TOT
, i.e. all the molecules are non-
phoshorylated at the beginning. We performed 10
4
Monte Carlo runs of SSA, with a time horizon of
500 seconds. The average number of reactions for
each SSA run was 9925. The whole computation took
more than 12 hours.
We then compared 3 methods of computing the
theoretical stationary distribution:
(1) We computed the eigenvalues and eigenvectors of
matrix
˜
G by means of Matlab’s eig routine. Ac-
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
686
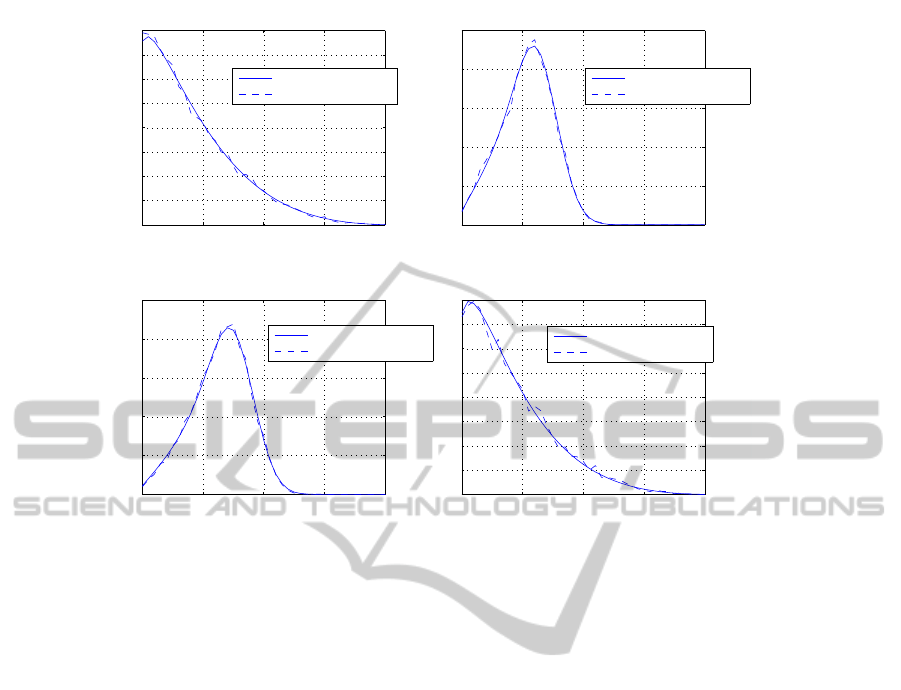
0 10 20 30 40
0
0.01
0.02
0.03
0.04
0.05
0.06
0.07
0.08
Stationary Distribution, Species A
(0)
Theoretical Distribution
Statistical Distribution
0 10 20 30 40
0
0.02
0.04
0.06
0.08
0.1
Stationary Distribution, Species A
(1)
Theoretical Distribution
Statistical Distribution
0 10 20 30 40
0
0.02
0.04
0.06
0.08
0.1
Stationary Distribution, Species A
(2)
Theoretical Distribution
Statistical Distribution
0 10 20 30 40
0
0.01
0.02
0.03
0.04
0.05
0.06
0.07
0.08
Stationary Distribution, Species A
(3)
Theoretical Distribution
Statistical Distribution
Figure 3: Steady-state marginal distributions. The solid line is the theoretical distribution computed explicitly according to
the methods illustrated in the paper. The dashed line is the statistical distribution provided by 5,000 Monte Carlo runs of the
Gillespie Algorithm.
cording to Proposition 2, the stationary distribu-
tion is the right eigenvector corresponding to the
zero eigenvalue of
˜
G. The procedure took 2785
seconds.
(2) According to Proposition 3, we computed
˜
P
ss
by
means of an ad hoc implementation of the Gaus-
sian Elimination method to compute the solution
of
˜
G
˜
P = 0. Although it is very accurate, this
method was outperformed (in terms of time com-
plexity) by method (1) and the elapsed time is
comparable to the Gillespie method.
(3) We computed the matrix exponential e
˜
G
, by
means of the Matlab routine expm, and we con-
sidered as initial condition
˜
P (0) the probability
vector with value equal to 1 for the component re-
ferred to the state n(0) and 0 elsewhere. We then
computed the steady-state distribution by means
of a fixed-point iteration for the recursion p
k+1
=
e
˜
G
p
k
, initialized with p
0
=
˜
P (0), which is the ex-
act discretization of the CME with unitary sam-
pling time, and where we used the stopping con-
dition kp
k+1
− p
k
k ≤ 10
−6
. Most of the time in
this case is spent in computing the matrix expo-
nential, computed in 357 seconds. The fixed point
iteration took 88 seconds to reach the steady state.
The methods above return the same distribution,
with a relative difference lower than 10
−6
(in norm).
The plots in Fig. 3 show the agreement between the
statistical estimation of the steady-state solution and
the theoretical stationary distribution. Due to the sav-
ing in terms of computational resources, the explicit
computation is preferable.
5 CONCLUSIONS
In this work we illustrated the application of some re-
sults about the efficient computation of the stationary
distribution of the Chemical Master Equation in the
biochemical framework of multisite phosphorylation.
Based on a recently developed representation scheme,
it is shown how the proposed approach results to be
accurate and with limited computational burden, es-
pecially with respect to the extensive use of statistical
Monte Carlo methods.
REFERENCES
Bazzani, A., Castellani, G. C., Giampieri, E., Remondini,
D., and Cooper, L. N. (2012). Bistability in the chem-
ical master equation for dual phosphorylation cycles.
The Journal of Chemical Physics, 136(23):235102.
Borri, A., Carravetta, F., Mavelli, G., and Palumbo, P.
(2013). Some results on the structural properties and
ChemicalMasterEquations-AMathematicalSchemefortheMulti-sitePhosphorylationCase
687

the solution of the chemical master equation. In Pro-
ceedings of the 2013 American Control Conference
(ACC 2013), Washington, DC, USA.
Bullo, F., Cortes, J., and Martinez, S. (2009). Distributed
Control of Robotic Networks. Applied Mathematics
Series. Princeton.
Carravetta, F. (2011). 2-d-recursive modelling of homoge-
neous discrete gaussian markov fields. IEEE Transac-
tions on Automatic Control, 56(5):1198–1203.
Castellani, G. C., Bazzani, A., and Cooper, L. N. (2009).
Toward a microscopic model of bidirectional synaptic
plasticity. Proceedings of the National Academy of
Sciences, 106(33):14091–14095.
Castellani, G. C., Quinlan, E. M., Cooper, L. N., and Shou-
val, H. Z. (2001). A biophysical model of bidirectional
synaptic plasticity: Dependence on ampa and nmda
receptors. Proceedings of the National Academy of
Sciences, 98(22):12772–12777.
Farina, L. and Rinaldi, S. (2000). Positive Linear Systems:
Theory and Applications. Pure and Applied Mathe-
matics Series. John Wiley & Sons, Inc.
Gillespie, D. T. (1977). Exact stochastic simulation of
coupled chemical reactions. The Journal of Physical
Chemistry, 81(25):2340–2361.
Mettetal, J. T. and van Oudenaarden, A. (2007). Necessary
noise. Science, 317(5837):463–464.
Michaelis, L. and Menten, M. (1913). Kinetics of invertase
action. Biochem. Z, 49:333–369.
Munsky, B. and Khammash, M. (2008). The finite state pro-
jection approach for the analysis of stochastic noise in
gene networks. IEEE Transactions on Automatic Con-
trol, 53(Special Issue on Systems Biology):201–214.
Qu, Z., Weiss, J. N., and MacLellan, W. R. (2003). Reg-
ulation of the mammalian cell cycle: a model of the
g1-to-s transition. American Journal of Physiology -
Cell Physiology, 284(2):C349–C364.
Ullah, M. and Wolkenhauer, O. (2011). Stochastic Ap-
proaches for Systems Biology. Springer.
Van Kampen, N. G. (2007). Stochastic Processes in Physics
and Chemistry, Third Edition. North Holland.
Whitlock, J. R., Heynen, A. J., Shuler, M. G., and Bear,
M. F. (2006). Learning induces long-term potentia-
tion in the hippocampus. Science (New York, N.Y.),
313(5790):1093–1097.
SIMULTECH2013-3rdInternationalConferenceonSimulationandModelingMethodologies,Technologiesand
Applications
688
