
Title-based Approach to Relation Discovery from Wikipedia
Rim Zarrad
1
, Narjes Doggaz
2
and Ezzeddine Zagrouba
1
1
RIADI Laboratory, Team of Research SIIVA, University of Tunis El Manar, Tunis, Tunisia
2
URPAH Laboratory, Faculty of Sciences of Tunisia, University of Tunis El Manar, Tunis, Tunisia
Keywords: Relation Extraction, Title, Ontology, Wikipedia.
Abstract: With the advent of the Web and the explosion of available textual data, the field of domain ontology
engineering has gained more and more importance. The last decade, several successful tools for
automatically harvesting knowledge from web data have been developed, but the extraction of taxonomic
and non taxonomic ontological relationships is still far from being fully solved. This paper describes a new
approach which extracts ontological relations from Wikipedia. The non-taxonomic relations extraction
process is performed by analyzing the titles which appear in each document of the studied corpus. This
method is based on regular expressions which appear in titles and from which we can extract not only the
two arguments of the relationships but also the labels which describe the relations. The resulting set of
labels is used in order to retrieve new relations by analyzing the title hierarchy in each document. Other
relations can be extracted from titles and subtitles containing only one term. An enrichment step is also
applied by considering each term which appears as a relation argument of the extracted links in order to
discover new concepts and new relations. The experiments have been performed on French Wikipedia
articles related to the medical field. The precision and recall values are encouraging and seem to validate
our approach.
1 INTRODUCTION
The exponential growth of the web contents has
transformed it into a universal information resource.
The need for developing methods allowing the
automation of processes like searching, retrieving
and maintaining information from Web documents
is obvious. In recent years, the field of ontology
learning and knowledge representation has attracted
a lot of attention, resulting in a wide variety of
approaches. Acquiring domain knowledge for
building ontologies is a difficult and time consuming
task, and would profit from a maximum level of
automation. Consequently, a significant number of
ontology learning tools and frameworks has been
developed aiming at the automatic or semi-
automatic ontology learning from structured,
unstructured or semi-structured documents.
There are currently three main paradigms
exploited to learn ontology from textual data. The
majority of these methods are based on the
techniques of natural language processing. The first
one focuses on the distribution of the linguistic units.
It focuses on studying the co-occurrence
distributions of terms in order to calculate a
semantic distance between the concepts represented
by those terms. Harris’ hypothesis (Harris, 1954),
which is the basis of word space models, states that
words that occur in similar contexts often share
related meaning. Although they are robust and do
not require preliminary knowledge on the field,
these methods disregard the context of sentences
which is necessary to have a precise interpretation of
the semantic classes, they are not adapted to have a
precise analysis of the corpus. The second paradigm
is based on syntactical process of the corpus (Faure
and Poibeau, 2000, Liu et al., 2005). It focuses on
the properties of the language to extract the
relationships between the ontology concepts. These
methods do not consider the corpus in a
comprehensive manner but locally. However, it is
not reasonable to specify a syntactical approach for
each new field of study. The third paradigm is based
on lexico-syntactic patterns (Morin, 1999, Ciarmita
et al., 2005; Snow et al., 2005). The user defines a
set of lexico-syntactic patterns (rules describing a
formed regular expression of words and grammatical
categories corresponding to the syntactic forms of
70
Zarrad R., Doggaz N. and Zagrouba E..
Title-based Approach to Relation Discovery from Wikipedia.
DOI: 10.5220/0004547400700080
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development (KEOD-2013), pages 70-80
ISBN: 978-989-8565-81-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

the relation and its arguments). These patterns
characterize the semantics of the relation. Parsing
and pattern matching methods were used effectively
in many approaches. Although the number of
extracted relations obtained when applying these
patterns would be very large, the main problem is
the complexity and of the diversity of the patterns
which can express the same relation.
The traditional methods for ontology learning
often privilege the analysis of the text itself. The
analysis of Web documents structure in order to
learn ontology components is a rather young field of
research. Most of the related works exploits HTML
tags for the analysis by building the explicit DOM
(Document Object Model) tree. The structure of a
HTML document may be considered as a hierarchy
where each document may have sections with
corresponding headings. A novel method is
proposed in (Pembe and Tunga, 2007) to solve the
problem of heading hierarchy identification for
HTML documents using a rule based approach and
DOM tree analysis. In this paper, we propose an
approach which extracts taxonomic and non-
taxonomic relationships between domain concepts.
Our approach explores and analyzes the title
hierarchy in each document in order to extract non-
taxonomic links. The title analysis method is
performed in three steps. Firstly, we focus on the
titles and subtitles composed by only one term to
extract relations between these terms. In the second
step, we check if the title structure corresponds to a
specific pattern that we have defined. This method
extracts not only non-taxonomic relations but also
corresponding labels which has been identified as
one of the most difficult problem for ontology
learning. In the last step, we use the discovered
labels in order to extract other relations.
Experiments are then conducted on a French corpus
collected from Wikipedia entries and related to the
medical field.
The rest of this paper is organized as follows. In
section 2, we present some useful definitions. A
variety of methods for relationships learning are
described in section 3. The general approach is
shown in section 3. In section 4, we describe the title
analysis method which extracts relations between
field concepts. The ontology enrichment process is
described in the section 5. We give details on the
used corpus and we give some experimental results
in section 6. Finally, we give our conclusion and we
present some perspectives.
2 PRELIMINARIES
In this section, we present some useful definitions
that we will use along our paper. These definitions
can vary according to theories or authors.
2.1 Concept versus Term
A concept represents a class of physical or abstract
objects. It is usually defined by a set of properties
that are both necessary and sufficient for belonging
to the class. Example: car, house, cat…
A term is a noun or compound word used in a
specific context. Several terms can denote the same
concept (Gomez and Benjamins, 1999). For
example, the concept "car" can be designated in the
text by: car, automobile, motor car, vehicle…
2.2 Taxonomic Relations
The taxonomic relation corresponds to the
hierarchical relation between two concepts (Guarino
and Welty, 2001). It is also called
hyponym/hypernym relation: hyponym is a noun
phrase whose semantic field is included within that
of another noun phrase, its hypernym. For example,
school is a kind of educational institution, so that a
taxonomic relation can be established between
school and educational institution.
2.3 Non-Taxonomic Relations
A non-taxonomic relation linking two concepts A et
B, also called functional relation, represents an
interaction between A and B (Gómez-Pérez et al.,
2000). In other words, the two concepts A et B are
linked by a non-taxonomic relation if A is
semantically related to B. These relations can be
active/passive relations, causal relations, locative
relations…
A label is generally assigned to a non-taxonomic
relation. Its role is to describe the relation between
the two concepts.
3 RELATIONS LEARNING
METHODS: STATE
OF THE ART
Ontology learning techniques focus on the extraction
of ontology elements such as concepts, instances or
relations. Learning taxonomic and non-taxonomic
relations between domain concepts is a crucial
Title-basedApproachtoRelationDiscoveryfromWikipedia
71

component in the ontology learning process. The
taxonomic relationships in ontologies are used to
organize concepts hierarchically whereas the non-
taxonomic ones describe other types of relations.
Several ontology learning tools have been developed
aiming to extract relationships from different kinds
of documents.
3.1 Taxonomic Relations Learning
The methods which deal with taxonomic relations
extraction focus generally on the analysis of the text
itself (Kermanidis and Fakotakis, 2007). Patterns
matching is one of the most used techniques to build
a taxonomy of concepts. For example, the approach
described in (Snow et al., 2005) learns syntactic
patterns for automatic hypernym discovery. It
extracts automatically hyponym (is-a) relations from
text using dependency paths and WordNet. Barbu et
Poesio describe a novel method which uses patterns
to build taxonomies of concepts from raw Wikipedia
text (Barbu and Poesio, 2009). The authors assume
that the concepts which are classified under the same
node in a taxonomy are described in a comparable
way in Wikipedia. Concepts belonging to six
taxonomies extracted from WordNet are mapped
onto Wikipedia pages and the lexico-syntactic
patterns describing semantic structures are
automatically learnt. Usually the results achieved
were promising. The reason behind this promising
result is that when applying patterns to the free text
the number of extracted semantic relations would be
very large. Nevertheless, the selection of patterns
should be done with caution because they should be
general enough so that they can give better
performance. The used language has also a great
impact on the ability of defining patterns and
generalizing them.
The analysis of documents structure in order to
learn ontology components is a rather young field of
research. Indeed, when a human reader tries to
understand the contents of a document, his or her
attention is focused on some particular elements
(Title, emphasized words,…) which are usually
more important than the rest of the document. The
structure analysis technique is used especially when
dealing with semi-structured data. In this context, a
study (Laurens, 2006) was made on a corpus of
XML documents to build a taxonomy of concepts.
This approach exploits only the visual structure of
the text (style, bold characters, underlined,
framing…) by making hierarchies of the text
elements according to their visual structure. The
intervention of the expert is necessary to validate the
field concepts. The authors in (Paukkeri et al., 2012)
propose also a method based on the document
structure for learning taxonomy from a set of
documents. Each document focuses on a domain
concept. Three different feature extraction
approaches are compared in this study in order to
reduce input data dimensionality by collecting the
relevant information and removing redundancies.
The first approach uses a combination of heuristic
criteria exploiting document structure (titles,
emphasized words, the first and the last part of a
document) by means of fuzzy logic. The second is a
language-independent approach based on statistical
stemming and keyphrase extraction. The third
approach is the traditional tf-idf weighting scheme
with commonly used rule based stemming. The Self-
Organizing Map, which is an artificial neural
network that orders data using unsupervised
learning, is then used to create an ordered space of
the concept vectors. We note that the use of
document structure in this method is applied only in
concept extraction phase and do not intervene in the
relation learning process. Sumida et Torisawa
(Sumida and Torisawa, 2008) propose to extract
hyponymy relations using the hierarchical layouts
from Wikipedia. Their method extracts more than
1.4 × 106 hyponymy relations from the Japanese
version of Wikipedia with a precision value equal to
75.3%. It uses also a machine learning technique,
pattern matching and other existing methods for
extracting relations from definition sentences and
category pages. However, we note that Wikipedia
categories hierarchy contains duplication and
sometimes it is inconsistent compared to other
manually created hierarchies like WordNet.
3.2 Non-taxonomic Relations Learning
In regards to ontology extraction, the identification
and labelling of non-taxonomic relations are
considered most challenging (Kavalec and Spyns,
2005). The works on non-taxonomic relations
discovery must not only extract relations between
concepts but also enable to label these relationships
in order to describe the relations.
The patterns and association rules are widely
used in non-taxonomic relations learning. For
example, Liu et al. (Liu et al., 2005) combine Hearst
patterns, head nouns, subsumption and co-
occurrence analysis in their approach towards
ontology extension. Their method is capable of
identifying hierarchical and unlabelled non-
hierarchical relations. In (Maedche et al., 2002), the
association rules are used to discover non-taxonomic
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
72

relations without labelling them further. This method
also covers the handling of relations between
instances of the same concept. Other non-taxonomic
relations learning systems are based on external
sources such as WordNet in addition to the above
approaches (patterns and association rules) in order
to extract relevant relations. For example, the work
presented in (Ruiz-casado et al., 2008) describes a
new procedure for the automatic semantic
annotation of the Wikipedia. It focuses on the
automatic association of Wikipedia entries with
nodes in WordNet. The proposed approach
combines linguistic processing, word sense
disambiguation and relation extraction techniques in
order to generate automatically patterns for
extracting taxonomic and non taxonomic relations.
Compared to extensive works on relations
learning, little attention has been given on labelling
of non taxonomical relations. Most of these works
focuses on the verbs in order to extract the labelled
relationships between terms which occur with these
verbs. Sanchez and Moreno (Sanchez and Moreno,
2008) start the process of learning non-taxonomic
relationships with the extraction of verbs from
sentences that contain domain concepts and
hyponyms of domain concepts. Those verbs are used
to retrieve and select related concepts. The approach
heavily depends on querying web search engines,
which provide suggestions for new concepts as well
as the verbs for relationship labelling. In contrast to
this approach, the method presented in
(Weichselbraun et al., 2009) relies exclusively on a
body of text to label unknown relations between
concepts. In order to retrieve the relation type, the
authors use machine learning techniques to compile
a knowledge base of verb vectors from known
relations and evaluate the method’s usefulness in
labelling unknown link types. More recently, Punuru
and Chen propose the VF*ICF metric for measuring
the importance of a verb as a representative relation
label, in the same spirit as the tf*idf measure in IR
(Punuru and Chen, 2012). Texts from two domains,
the electronic voting domain texts and the tenders
and mergers domain texts are used to compare the
method with one of the existing approaches.
To the best of our knowledge, the works on
labelled relations discovery are generally based on
syntactical analysis and especially on the verbs
appearances in sentences. In the rest of the paper, we
present our method which extract taxonomic and
non-taxonomic relations and gives in most cases the
corresponding labels. The non-taxonomic relations
discovery is realized by analyzing the hierarchy of
titles in each Wikipedia article.
4 GENERAL APPROACH
We present in figure 1 the general approach for
relations extraction and ontology enrichment. In our
approach, we use Wikipedia entries as a corpus and
several natural language processing tools to analyze
the collected corpus. The corpus pre-processor is
performed using the tree Tagger tool and the
HtmlParser 1.6 necessary for the extraction and the
text processing of a web corpus. Two stop-lists are
also used to eliminate uninteresting words and titles.
After the pre-processing step, we proceed to extract
the main concepts of the studied field by applying
the method proposed in (Zarrad et al., 2012a; Zarrad
et al., 2012b) which uses information on the
document structure to extract relevant information.
The taxonomic relations are discovered using
syntactical method and patterns matching technique.
In our approach, we propose also a method which
focuses on the hierarchy of titles in each Wikipedia
article to extract non-taxonomic relations. This
method is established in three steps: reduction phase,
titles pattern phase and three-level analysis.
In this section, we describe the corpus pre-
processing step. We present then our methodology
for discovering domain concepts and taxonomic
relations among the extracted concepts. The non-
taxonomic relations discovering approach and the
ontology enrichment process will be detailed in the
following sections.
In order to build domain ontology, we have
divided the learning process into several steps:
4.1 Corpus Pre-processing
The approaches of ontologies learning from text are
generally based on a corpus of texts. This corpus
must be representative of the field for which we try
to build the ontology. The corpus pre-processing
step is performed using several natural language
processing tools (NLP). In our approach, this phase
is realized in three steps:
Part-of-speech tagging. We use the Tree Tagger
tool (Schmid, 1994) in order to associate to each
instance of a word its grammatical category (noun,
verb,…) and its canonical form.
Parsing. We use HtmlParser 1.6 which analyzes
and extracts data from the tags of the HTML
documents.
Removing stop-words and stop-titles. A general
stop-word set is used to locate the stop-words in
the corpus (articles, prepositions, conjunctions…).
Another stop-title set is used in order to eliminate
Title-basedApproachtoRelationDiscoveryfromWikipedia
73
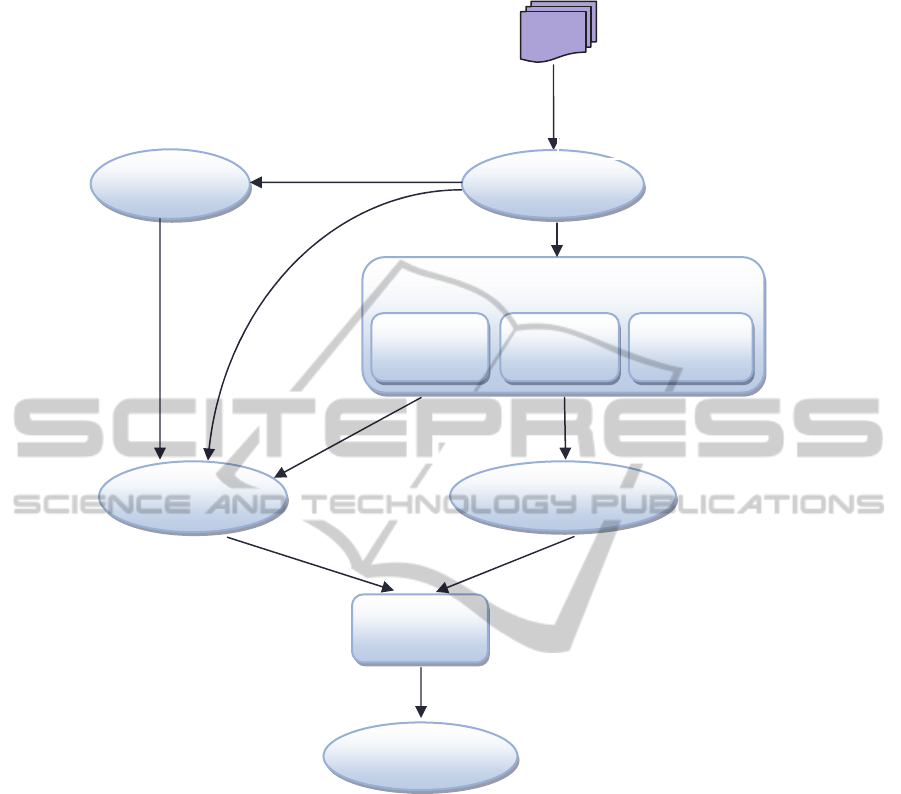
Figure 1: General approach.
titles which occur frequently in general structured
texts and especially in Wikipedia articles
(references, external link…).
4.2 Concepts Extraction
Our approach rests on the extraction of the candidate
terms (CT) from the studied corpus. They are
linguistic units which qualify an object or a concept
of the real-world. We extract two types of terms
according to their canonical syntactic structures.
─ Class 1: terms composed by only one word, they
can be either a “Name” or a “Named_Entity”
─ Class 2: terms containing two words: they have
as syntactic structure the sequence “Name
Adjective” or “Named_Entity Adjective”.
After extracting the candidate terms from the
corpus, they will be filtered by analyzing the
documents’ structure in order to extract the main
concepts of a given field. Indeed, the material form
of the documents provides interesting information
on the semantics contained in the texts. We have
presented in (Zarrad et al., 2012) an approach which
filter the extracted CT using a new measure denoted
CR-ICF.
The CR factor is based on the occurrences of the
CT in the titles, the links and the used styles in the
documents, whereas the ICF one is based on the
occurrences of the CT in other corpora in order to
check if the CT is a general term or it is specific to
the studied field.
Title Analysis
Analysis
Title
Reduction
Titles
Patterns
Preprocessed
Corpus
Wikipedia articles
Concepts
Taxonomic
Relations
Non-Taxonomic
Relations
Syntactical Process
Patterns Matching
Three-level
Analysis
Tree Tagger
HTML Parser
Stop sets
Enrichment
Process
New concepts
and new relations
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
74

4.3 Taxonomic Relationships Retrieval
The taxonomic relations classify the concepts from
the most general to the most specific ones. We
propose in this paper to use a syntactical approach
and apply patterns matching method in order to
extract taxonomic links between concepts. Indeed,
the results obtained by these methods are usually
relevant and have high precisions.
4.3.1 Syntactical Process
This approach focuses on the syntactic structure of
the domain concepts in order to discover taxonomic
relations between them. When analyzing the two
classes of concepts, we can easily deduce that a
taxonomic relationship between the two types of
concepts can be extracted. Indeed, an adjective is a
describing word which modifies the word that
precedes by describing it or making it more specific,
thus the sequence “Name Adjective” (respectively
“Named_Entity Adjective”) is a specification of
“Name (resp. “Named_Entity”). Our approach
considers a concept C
2
belonging to the second class
as a sub-concept of another concept C
1
which
belongs to the first class if it is composed by the
concept C
1
followed by an adjective.
4.3.2 Pattern Matching Method
We use the lexico-syntactic patterns in order to
detect the taxonomic links between the domain
concepts. They are rules describing a regular
expression of words and grammatical categories
corresponding to the syntactic forms of the relation
and its arguments. These patterns characterize the
semantics of the relation. In our case, this method
extracts the syntactic contexts which "mark" the
hyperonymy link between a potential couples
(Term
1
,Term
2
) where the term Term
1
is more general
than the term Term
2
. We use the patterns defined by
Marshman which are specific to hyperonymy
relation in French language (Marshman, 2008). We
have also extended this list by other patterns defined
manually:
Term such as Term
1
,… Term
i
(Term tel que Term
1
,… Term
i
)
Term particularly Term
1
,… Term
i
(Term en particulier Term
1
,… Term
i
)
Term especially Term
1
,… Term
i
(Term notamment Term
1
,… Term
i
)
Although, these patterns describe taxonomic
relations, they give in some cases unsatisfactory
results. To improve the results, we set up a
hypothesis in order to eliminate cases that we judge
invalids. In fact, if Term is preceded by the
preposition "of" or "of this", then no hierarchical
relationship can be established between Term and
each term Term
i.
To illustrate this idea, let consider
the following sentence:
Transient contamination of the blood is
bacteremia
In our approach, we consider only the concepts
having as syntactic structure: noun or noun followed
by an adjective. If we check the pattern “Term
1
is a
Term
2
” defined by Marshman for the extraction of
hierarchical relation, we conclude that “blood” is a
sub-class of “bacterimia”.
This invalid result is due to the use of the
preposition “of”. Indeed, the verb “to be” links
actually the two terms “transient contamination” and
“bacterimia”. Our approach extracts in this case a
hierarchical relationship between these two terms.
The real problem when applying the syntactical
approach is the weak production. In fact, this
approach takes the list of concepts as input, so that
the number of retrieved relations depends on the
number of concepts from each class. We note also
that despite the pattern matching method extracts a
large set of relations, the number of extracted
taxonomic links is still low compared to those
generated by the statistic approaches. To improve
the production when extracting the taxonomic links,
we propose to extend the linguistic and the lexico-
syntactic patterns phases by considering the
structure and specially the titles of the documents.
5 RELATIONS EXTRACTION
APPROACH USING TITLES
In the ontology learning process, the discovery, and
possibly also labeling, of non-taxonomic
relationships among concepts has been identified as
one of the most difficult problems. Thus, it would be
efficient to automate or semi-automate the
acquisition of non-taxonomic relations among
domain concepts. In our approach, we consider the
titles of the documents for the extraction of the
relations between domain concepts. The titles are
extracted by analyzing the HTML tags of each
document. According to (Jacques and Rebeyrolle,
2006), “The nested titles of sub-sections belonging
to a given section reflects the nested relations
existing between these sections”. The text can be
then considered not like a linear succession of
blocks, but like a structure of elements of high level
Title-basedApproachtoRelationDiscoveryfromWikipedia
75

which include other elements.
Let TermSet(T) be the set of terms belonging to a
title T:
,
,…
|
|
Moreover, we use a tree structure in order to
represent the hierarchy of titles in each document.
Definition
For each Wikipedia article A, Let Ttree(A) be the
tree (Root, Nodes, Edges) defined as follows:
Root: the title of the Wikipedia article A.
Nodes: the set of all titles and subtites that belong to
the Wikipedia article A.
Edges: each edge (T
s
,T
f
) links two titles T
s
and T
f
belonging to the same Wikipedia article A where T
s
is a subtitle of T
f
Notations
In the rest of the paper, we note T
s
and T
f
two titles
belonging to the same Wikipedia article A where T
s
is a subtitle of T
f
.
Each taxonomic relation is described by the two
arguments of the relation and we note it:
,
.
The non-taxonomic relation is described by the
two arguments and the label of the relation and we
note it:
,
,
Our approach is established in three phases:
Title Reduction step
Title Pattern step
Three-level step
5.1 Reduction Phase
This method operates on titles and their sub-titles
and generates relations among the terms they
contain. To produce good results, we focus on titles
containing only one term. Indeed, when the number
of terms in titles is high, it is difficult to extract
relations between the terms belonging to these titles.
Our method operates as follows: if S
1
is the only
term which appears in the title T
f
and S
2
is also the
only term belonging to the title T
s
(T
s
is a subtitle of
T
f
) then a relationship can be established between S
1
and S
2
. More formally:
∀
,
uEdges
|
|
1
Establishrelation
,
Although, the retrieved relationships are sometimes
taxonomic ones, we notice that in most cases, they
express other types of relations. As an example, the
relation linking the pair of terms (tooth, dental
anomaly) which is extracted when applying this
method on a Wikipedia article is a non-taxonomic
relation. The identification of the type of the
extracted relationships is performed by a domain
expert.
5.2 Title Pattern Phase
In this step, we propose a method which analyzes
each title in each document of the domain corpus in
order to discover non-taxonomic relationships. To
achieve this goal, we define manually regular
expressions which are very used in French language
and from which we can extract not only the two
arguments of the relationships but also the labels
which describe the relations.
Our method takes as input two titles T
s
and T
f
of
the same Wikipedia article, where T
s
is a subtitle of
T
f
. If the title T
f
has as syntactic structure the
sequence:
Term of Term
1,
Term
2
,… and Term
n
and T
s
is a Term Term’, we establish n relationships
labelled Term. The two arguments of each extracted
relation are Term’ and Term
i
where 1 i n.
We note that if the label of the extracted relation
corresponds to “type” or “kind”, the relation
becomes a taxonomic one. Indeed, the term Term’
belonging to the sub-title T
j
becomes a "type" of that
(or those) appearing after the preposition of in the
original title T
i
. In the other cases, the relation is
classified as a non-taxonomic one and will be
automatically labelled.
More formally, our method operates as follows:
Let prepSet be the set:
"de","′",""
//
"of"
∀Edgesu
,
,
,…
|
|
1
1
if
"type"
"kind"
establishtaxonomic‐relation
,
else
establishnon‐taxonomic‐relation
,
,
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
76

As an example, we extract from a Wikipedia
document the two following titles:
Disease of immunity «Maladie de l’immunité»
Immune deficiency «Déficit immunitaire»
Our approach retrieves a relationship labeled
“disease” between the two concepts “immunity” and
“immune deficiency”. This relationship is valid and
relevant in the medical field since immune
deficiency is a disease of the immune system.
Figure 2: Example of extracted relations.
Notation
We note LabSet the set of labels discovered in the
title pattern step extended manually by other labels
which we judge relevant to the studied field such as
(symptom, cause, treatment, diagnostic,…).
5.3 Three-Levels Analysis
This method
analyzes the title hierarchy in each
document in order to learn labeled relations. It takes
as input the set of labels learned when applying the
title patterns step (Labset). We can use the set of
labels in order to extract other couples which convey
the relation. This is realized by analyzing three
levels of the titles hierarchy. Indeed, if an extracted
label lab constitutes the only term of the medium
level title, than we extract a relation labeled lab
between the terms belonging to the title of the higher
level and those belonging to the lower one.
More Formally:
∀Edgesu
,
Edgesu′
,
|
||
|
||1
"
"
"type"
"kind"
establishtaxonomic‐relation
,
else
establishNon‐taxonomic‐relation
,
,
For example, we extract from a Wikipedia document
the titles (bronchiolitis, treatment, kinesitherapy)
where kinesitherapy is a subtitle of treatment and
treatment is a subtitle of bronchiolitis. Since
treatment belongs to the set of labels, we extract a
relation labelled treatment between the two concepts
bronchiolitis and kinesitherapy.
6 ONTOLOGY ENRICHMENT
As shown in figure 1, the ontology enrichment
process takes as input the extracted relations when
applying the relationships discovery methods. It
aims to retrieve new concepts and new relations and
integrates them into the obtained ontology.
6.1 New Concepts Discovery
Among the proposed methods to discover taxonomic
and non-taxonomic relations, we notice that only the
syntactical one takes the list of concepts as input,
whereas all the other methods extract relations
without considering the set of domain concepts. The
syntactical process establishes taxonomic relations
between the two classes of concepts. Thus, each
extracted relation has as arguments two concepts
belonging to the set of concepts already found. This
is not the case when applying the other methods.
Indeed, all the relations obtained when projecting
the lexico-syntactic patterns or by analyzing the
titles of Wikipedia articles, do not link necessary
two domain concepts. Thus, the arguments of these
relations can be terms which not belong necessarily
to the set of concepts already found. Since, each of
these relations where validated by an expert of the
studied field, we propose to add each term which
appears as a relation argument. As an example,
when applying the patterns and titles analysis
methods on a corpus from Wikipedia entries related
to the medical field, we obtain a taxonomic
relationship among the pair of terms (specialty,
oncology). Although, these two terms were not
retrieved in the concepts selection step, we conclude
that specialty and oncology can be considered as
main concepts of the studied field and consequently
they are added to the list of domain concepts.
6.2 New Relations Extraction
Since we have extracted new concepts, we conclude
intuitively that other relations could be retrieved to
enrich the ontology. These relations are discovered
using the linguistic method which extracts on the
one hand new taxonomic relationships among these
concepts and on the other hand new taxonomic
Immune
deficiency
disease Immunity
Title-basedApproachtoRelationDiscoveryfromWikipedia
77
relationships linking them to the other concepts. We
enrich the list of relations by adding each new
taxonomic relation obtained by applying the
linguistic method.
7 EXPERIMENTS AND RESULTS
Wikipedia is a collaboratively edited, multilingual,
free Internet encyclopedia. It is one of the most
visited sites on the web, outstrips all other
encyclopedias in size and coverage (Medelyan et al.,
2009). It is also an important semi-structured
knowledge source which contains a rich body of
lexical semantic information that has been used to
extract relationships (Zesch et al., 2008). The
Wikipedia articles are highly structured with
headings which are conform to specific guidelines.
In order to evaluate our approach, we have then used
a corpus of 90 Wikipedia articles related to the
medical field that we have collected randomly. The
studied corpus contains 3514 titles. However, about
one-third of these titles are not considered as they
belong to the stop-title set.
After pre-processing the collected corpus, and
applying the concept extraction approach, we obtain
as result a total number of 617 concepts: 524
concepts belonging to the class 1 and 93 concepts
belong to the class 2. The same corpus is used in
order to discover relationships between these
concepts. When running the developed application
described in section 4 and 5, we obtain 1079
relationships among the extracted concepts. To
evaluate the results, a domain expert checks if each
retrieved relationship is relevant in the medical field.
We use the same definitions of precision and recall
known in IR (Powers, 2007).
∩
∩
The number of the extracted relations and the
precision values obtained while applying each
method is given in the table 1. As we can note, the
number of the extracted relationships using the
syntactical process is low towards those obtained by
applying patterns matching and by analyzing the
titles of each Wikipedia article. Indeed, the first
method extracts relations linking concepts belonging
to the two classes of concepts. The number of
retrieved relations is then dependant of the number
of extracted concepts from each class. We note that
although this method gives a weak production, it
achieves a high precision value equals to 97.36%.
The patterns projection method gives the lower
precision value which is equal to 75.7%.
Nevertheless, it uses both patterns list presented by
Marshman (Marshman, 2008) and other patterns that
we have defined manually. Thus the number of
extracted relations reaches 424 which is relatively
high. Moreover, it is remarkable to note that when
we use the patterns projection method without the
use of the patterns hypothesis described in the
section 4.2, we obtain 62.57% as precision value.
Table 1: Evaluation of the extracted relationships using
described methods.
Syntactical
Process
Patterns
Matching
Titles
Analysis
Extracted
Relations
Number
38 424 617
Correct
Relations
Number
37 321 525
Precision 97.36% 75.7% 85.09%
A total of 617 relations have been automatically
extracted from the Wikipedia entries using the title
analysis method that was described in the section 5.
The results obtained by each step of the title analysis
method are detailed in table 2. The precision value
obtained by the analysis method ranges from
83.84% and 96.22%.
The three steps (title reduction, title patterns and
three-level analysis) give taxonomic and mainly
non-taxonomic relations. Except for relations
obtained using the title reduction step which are
labelled by a field expert, those obtained by applying
the title patterns and three-level steps are labelled
automatically without the intervention of the expert.
When computing the recall number corresponding to
the title analysis method, we obtain as result 54.55%
for taxonomic relation retrieval and 68.42% for non-
taxonomic relation retrieval.
As described in the previous section, we proceed
to enrich our ontology by discovering new concepts
and new taxonomic relationships. As a result, we
obtain 397 new concepts belong to the class 1 and
311 concepts belong to second class. When
reapplying the syntactical method, we extract 226
new taxonomic relations. Among these links, 220
were validated by a domain expert which
corresponds to a precision value equal to 97.34%.
To the best of our knowledge, all the methods
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
78
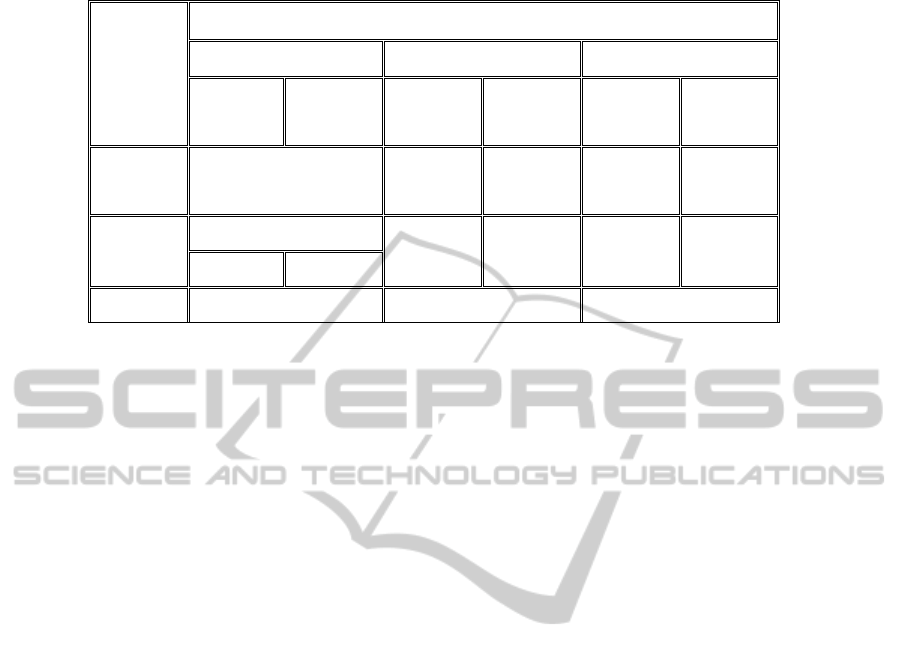
Table 2: Evaluation of the extracted relationships using the titles analysis method.
Titles analysis
Title reduction step Title pattern step Three-level step
Taxonomic
Relations
Non-
Taxonomic
Relations
Taxonomic
Relations
(type of)
Non-
Taxonomic
Relations
Taxonomic
Relations
(type of)
Non-
Taxonomic
Relations
Extracted
Relations
Number
526 18 36 1 36
Correct
Relations
Number
441
18 34 1 31
182 259
Precision 83.84% 96.22% 86.48%
which rely on document structure and especially on
titles hierarchy extract only taxonomic relations. For
example, the method presented by Sumida and
Torisawa (Sumida and Torisawa, 2008) uses
hierarchical layouts (headings, bullet list and ordered
list) in order to extract relations from the Japanese
version of Wikipedia. Although their method
retrieves a large set of relations, all the extracted
relations are taxonomic ones. In the same way, the
method presented by authors in (Paukkeri et al.,
2012) focuses on titles and emphasized words in
order to learn taxonomic relationships from a set of
HTML documents. Our method is not restricted to
taxonomic relations but also extracts labelled
non- taxonomic relationships from Wikipedia.
Moreover, the precisions values obtained by our
method are better than those found by (Paukkeri et
al., 2012; Laurens, 2006; Sumida and Torisawa,
2008).
8 CONCLUSIONS AND FUTURE
WORK
In this paper, we have presented a method which
extracts from Wikipedia entries taxonomic and non-
taxonomic relations. The taxonomic links are
retrieved by combining a syntactical method which
focuses on the syntactic structure of the domain
concepts and another method which applies patterns
on the studied corpus. The main contribution in this
paper is the title analysis method which extracts
non-taxonomic relations by analyzing the titles of
each document. This method is performed in three
steps: title reduction, title patterns, and three-level
analysis. The identification of the relations type is
committed by an expert when applying the title
reduction Step. In case of title patterns step, our
system detects automatically if the considered
relation is taxonomic or non-taxonomic by checking
the label of the relationship. The Three-Level step
takes as input the set of labels obtained by the Title-
Patterns step and gives as output a set of non-
taxonomic relations with the associated labels. An
enrichment step is also applied by considering each
term which appears as a relation argument of the
extracted links in order to discover new concepts
and new relations.
As future work we aim to:
1. Increase the corpus size in order to extract more
relationships. Since the field on which we work
is wide and contains a huge number of concepts,
we will probably obtain more interesting results
when the number of documents in the corpus is
high.
2. In the title reduction step, it would be interesting
to consider the titles containing more than one
term in order to extract other relations. Several
assumptions must be identified in order to detect
the relation type, the relation arguments and the
corresponding label in case of non-taxonomic
relation.
ACKNOWLEDGEMENTS
The authors are thankful to Dr. Imen Jmour for her
valuable and thoughtful feedback in the validation
task as an expert in the medical field.
REFERENCES
Barbu, E., Poesio, M., 2009. Unsupervised Knowledge
Title-basedApproachtoRelationDiscoveryfromWikipedia
79

Extraction for Taxonomies of Concepts from
Wikipedia. In: Proceedings of the International
Conference in Recent Advances in Natural Language
Processing, RANLP-2009. Bulgaria. pp. 28-32.
Ciarmita, M., Gangemi, A., Ratsch, E., Jasmin, S. Isabel,
R., 2005. Unsupervised Learning of Semantic
Relations between Concepts of Molecular Biology
Ontology. In: Proceedings of the 19th international
joint conference on Artificial intelligence, IJCAI'05.
pp. 659-664.
Faure, D., Poibeau, T., 2000. First experiences of using
semantic knowledge learned by ASIUM for
information extraction task using INTEX. In: ECAI
Workshop on Ontology Learning, ECAI’2000.
Germany.
Gomez P. A., Benjamins V.R., 1999. Overview of
Knowledge Sharing and Reuse Components:
Ontologies and Problem-Solving Methods. IJCAI and
the Scandinavian AI Societies. CEUR Workshop
Proceedings.
Gómez-Pérez, A., Moreno, A., Pazos, J., Sierra-Alonso,
A., 2000. Knowledge Maps: An essential technique for
conceptualization. In Data & Knowledge Engineering.
33(2). pp 169-190.
Guarino, N., Welty, C., 2001. Identity and Subsumption,
In The Semantics of Relationships: an
Interdisciplinary Perspective. R. Green, C.A. Bean, S.
Hyon Myseng (Eds). Kluwer. pp 111-126.
Harris, Z., 1954. Distributional structure. Word 10 (23).
pp. 146–162.
Jacques, M.P, Rebeyrolle, J., 2006. Titres et structuration
des documents. In: Actes International Symposium:
Discourse and Document, ISDD'06. France. pp. 01-12.
Kavalec, M. and Spyns, P., 2005. Ontology Learning from
Text, chapter A Study on Automated Relation
Labelling in Ontology Learning. IOS Press.
Amsterdam. pp 44–58.
Kermanidis, K., Fakotakis, N., 2007. One-sided Sampling
for Learning Taxonomic Relations in the Modern
Greek Economic Domain. In Proceedings of the 19
th
IEEE Tools with Artificial Intelligence, ICTAI, Vol.2.
pp. 354-361
Laurens, F., 2006. Construction d'une ontologie à partir
de textes en langage naturel. Master report. University
Paris7.
Liu, W., Weichselbraun, A., Scharl, A., Chang, E., 2005.
Semi-automatic ontology extension using spreading
activation. Journal of Universal Knowledge
Management. vol. 1. pp. 50–58.
Maedche, A., Pekar, V., Staab, S., 2002. Ontology
learning part one - on discoverying taxonomic
relations from the web. Web Intelligence. In Zhong,
N., Liu, J., and Yao, Y., editors. Springer. pp. 301–
322.
Marshman, E., 2008. Expressions of uncertainty in
candidate knowledge-rich contexts. Terminology. Vol.
14, Number 1. pp 124-151.
Medelyan, O., Milne, D., Legg, C., Witten, I. H., 2009.
Mining meaning from Wikipedia. International Journal
of Human-Computer Studies. vol. 9. pp 716-754.
Morin, E., 1999. Using Lexico-Syntactic Patterns to
Extract Semantic Relations between terms from Tech
nical Corpus. In: Proceedings of the 5
th
International
Congress on Terminology and Knowledge
Engineering, TKE'99. Austria. pp 268-278.
Paukkeri, M.S., García-Plaza, A.P., Fresno, V., Unanue,
R.M., Honkela, T., 2012. Learning a taxonomy from a
set of text documents. Journal of Appl. Soft Comput.
Elsevier Science Publishers B. V. Vol 12. pp 1138-
1148.
Pembe, F.C., Tunga, G., 2007. Heading-based sectional
hierarchy identification for HTML documents. 22nd
international symposium on Computer and
information sciences. pp 1-6.
Powers, D. M., 2007. Evaluation: From precision, recall
and f-factor to roc, informedness, markedness &
correlation. School of Informatics and Engineering.
Adelaide, Australia.
Punuru, J., Chen, J., 2012. Learning non-taxonomical
semantic relations from domain texts. Journal of
Intelligent Information Systems. vol. 38. pp 191-207.
Ruiz-casado, M., Alfonseca, E., Okumura, M., Castells,
P., 2008. Information Extraction and Semantic
Annotation of Wikipedia. Ontology Learning and
Population: Bridging the Gap between Text and
Knowledge. pp 145-169.
Sanchez, D., Moreno, A., 2008. Learning non-taxonomic
relationships from web documents for domain
ontology construction. Data and Knowledge
Engineering. vol. 64. pp 600–623.
Schmid, H., 1994. Probabilistic Part-of-Speech Tagging
Using Decision Trees. In: Proceedings of the
International Conference on New Methods in
Language Processing. Manchester, UK. pp. 44-49.
Snow, R., Jurafsky, D., Ng, A. Y., 2005. Learning
syntactic patterns for automatic hypernym discovery.
In: Nineteenth Annual Conference on Neural
Information Processing Systems, NIPS 2005.
Vancouver, Canada. pp 1297-1304.
Sumida, A., Torisawa, 2008. T. Hacking Wikipedia for
hyponymy relation acquisition. Proceedings of
IJCNLP. pp 883–888.
Weichselbraun, A., Wohlgenannt, G., Scharl, A.,
Granitzer, M., Neidhart, T. Juffinger, A., 2009.
Discovery and evaluation of non-taxonomic relations
in domain ontologies. International Journal of
Metadata, Semantics and Ontologies. Vol. 4. pp. 212-
222.
Zarrad, R., Doggaz, N. and Zagrouba, E. 2012a. Concepts
Extraction based on HTML Documents Structure. In
Proceedings of the 4th International Conference on
Agents and Artificial Intelligence, ICAART2012.
Vilamoura, Algarve, Portugal, pp. 503-506.
Zarrad, R., Doggaz, N., Zagrouba, E., 2012b. Toward a
Taxonomy of Concepts using Web Documents
Structure. In Proceedings of the 14
th
International
Conference on Information Integration and Web-based
Applications & Services, IIWAS2012. Bali, Indonesia.
pp. 303-312.
Zesch, T., Müller, C., Gurevych, I., 2008. Extracting
Lexical Semantic Knowledge from Wikipedia and
Wiktionary. In: Proceedings of the Sixth International
Conference on Language Resources and Evaluation,
LREC'08. Morocco.
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
80
