
A Linked Data Approach for Querying Heterogeneous Sources
Assisting Researchers in Finding Answers to Complex Clinical Questions
Nikolaos Matskanis
1
, Vassiliki Andronikou
2
, Philippe Massonet
1
,
Kostas Mourtzoukos
2
and Joseph Roumier
1
1
Centre d’ Excellence en Technologies de l’ Information et de la Communication (CETIC),
Rue des Frères Wright 29/3, Chareroi, B-6041, Belgium
2
Telecommunications Laboratory Department of Electrical & Computer Engineering,
National Technical University of Athens, 9 Heroon Polytechniou, 15773, Athens, Greece
Keywords: Semantic Search, Heterogeneous Data Sources Querying, Semantic Aggregation of Data.
Abstract: Clinical trials for drug repositioning aim at evaluating the effectiveness and safety of existing drugs as new
treatments. This involves managing and semantically correlating many interdependent parameters and
details in order to clearly identify the research question of the clinical trial. This work, which is carried out
within the PONTE (Efficient Patient Recruitment for Innovative Clinical Trials of Existing Drugs) project,
aims to improve the trial design process, by not only offering access to a variety of relevant data sources –
including, but not limited to, drug profiles, diseases and their mechanisms, genes and past trial results – but
also providing the ability to navigate through these sources, perform queries on them and intelligently fuse
the available information through semantic reasoning. This article describes our intention to consume and
aggregate information from Linked Data sources in order to produce answers for the clinical researcher’s
questions.
1 INTRODUCTION
Clinical research, including investigation of the test
of hypothesis, clinical trial design and study
conduction, comprises an expensive and very
important part of the R&D activities of the
pharmaceutical companies. Trials for drug
repositioning, which aim at testing established
compounds to new medical conditions, appear to
gain ground as their compounds have already been
found safe in clinical trials and sometimes are
already present in the market. Although the
uncertainty of safety is lower in such trials, still
problematic situations – for example an adverse
effect of a drug compound - present important risks
(Ashburn, 2004). In many cases evidence of such
issues already exists in published scientific literature
and other clinical information sources but is often
difficult to discover and/or correlate, given the
volume, variety and distributed nature of the
information required.
Scientific results from medical research,
including those from clinical trials, are gradually
being published as Linked Open Data (LOD) (Bizer,
2010). Although not many of the available clinical
data sources are available as Linked Data, there is a
good and growing representation that can be used
for the purposes of the PONTE project
(Chondrogiannis, 2011). This rapidly growing
collection of well-structured (using RDF and OWL)
and easily accessible (using HTTP) data (Berners-
Lee, 2006) contains clinical information about drugs
and their side effects, clinical trials, diseases and
their mechanisms (pathophysiology) and other
aspects of the domain. These data sources comprise
a valuable source for clinical research but more
importantly, because of their structure and links,
they can be used by search tools to produce search
results that are otherwise hard or impossible to come
across.
The idea of deploying semantic web technologies
for querying and creating links between data sources
for enabling automatic knowledge aggregation is an
area of scientific research that several projects are
exploring and developing technologies and tools for.
The Bio2RDF project has created a framework that
provides on demand data for mash-ups in the
bioinformatics domain (Belleau, 2008). There are
411
Matskanis N., Andronikou V., Massonet P., Mourtzoukos K. and Roumier J..
A Linked Data Approach for Querying Heterogeneous Sources - Assisting Researchers in Finding Answers to Complex Clinical Questions.
DOI: 10.5220/0004173004110414
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development (KEOD-2012), pages 411-414
ISBN: 978-989-8565-30-3
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)
projects that have investigated several techniques on
querying the Linked Data cloud (Bouquet, 2009),
(Hartig, 2012). The FedBench project (Schmidt,
2011) has produced a benchmark framework for
analysing the efficiency and performance of
different strategies for federated query processing on
semantic data. In this paper we will explain the
benefits of our semantic search approach that aims at
assisting the design of clinical trials on existing
drugs by discovering semantic associations within
the Linked Data sources.
2 INCORPORATION AND
AGGREGATION OF LINKED
DATA SOURCES
During clinical research different parameters about
drugs, diseases, trial targets (genes, proteins) are
assessed and carefully examined. This information is
dispersed in different data sources created
autonomously by often-unassociated institutional
bodies, then – some of it - republished as Linked
Data (Samwald, 2011). In order to effectively use
the knowledge existing within these sources we need
to combine the information available. For instance,
the information about the possible disease targets of
a given drug can be found in the DrugBank Linked
Data source. However, the information about the
genes associated with a specific disorder is found in
Diseasome Linked Data source.
Thanks to the
SPARQL endpoints we are able to collect the
information from all these sources. This would
otherwise be a quite difficult task with only the
upstream data sources, since we would have to
overcome a variety of problems, including the
database technology, the various structure logics and
the way information is represented.
Additionally we have chosen to use the
ontologies of some of the above data sources for
describing the query term, defining its domain (by
using a taxonomy graph) and the data aggregation
options for presenting the information. The domain
taxonomy graph is composed using Construct
queries on the ontology of the term, while the related
concepts graph is constructed by using the RDF
links and relations between the query term and
concepts or instances found in the other domain
ontologies. For reasons of availability and
performance of the system the ontologies are hosted
in an RDF repository, which is part of the Ontology
Management component (see figure 1) and are made
available to PONTE components through a
SPARQL endpoint inside a common and closed
network.
2.1 The Linked Data Application and
Query Engine
The Linked Data Application (LDApp) is part of a
software framework created in the context of
PONTE project (Chondrogiannis, 2011). Its primary
function is to assist the investigation on a drug, its
target, and study disorder. The LDApp is a web
application that dynamically generates a data
aggregation of information retrieved from the
PONTE platform – more specifically from the
Ontology Management component - and the Linked
Open Data Cloud. The LDApp uses the SPARQL
query language for RDF to retrieve RDF Graphs
from the Ontology Management component and to
query the Linked Data sources. In terms of the
LDApp interfacing with the other components of the
platform, the queries that are generated from these
components contain information that associates the
search term with the domain of the question. Using
this information, the main term or concept of the
query is linked with a concept from the respective
Ontology. The interactions between the LDApp, the
PONTE components and the Linked Data sources
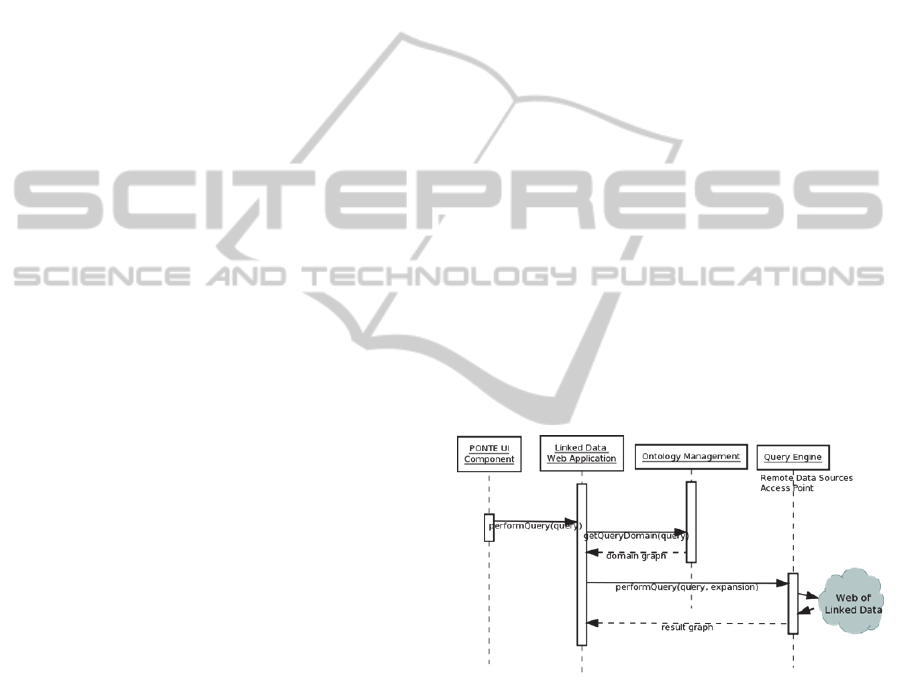
are presented in the sequence diagram in Figure 1.
Figure 1: Component Interactions for Question
Answering.
The LDApp requests from the RDF repository of
the Ontology Management component the related
concepts of the domain ontologies. It then constructs
a graph that is composed of concepts and instances
representing the taxonomy and relevant information
of the query’s main term. The graph concepts and
instances additionally contain relations and RDF
links to the web of Linked Data sources, which can
be used to execute queries across multiple data
sources. The graph is visualized by the LDApp and
can be used to assist the answering of research
KEOD2012-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
412

questions by providing options to expand or filter
the query results.
The LDApp can perform additional queries to
retrieve data for constructing a Linked Data mash-
up. These queries to Linked Data sources are
performed via the PONTE Query Engine. The Query
Engine is a SPARQL endpoint that is able to execute
queries across multiple data sources and for the
purposes of this prototype implementation, its
developments are based on the SQUIN library that
supports link traversal based query execution
(Hartig, 2009).
Figure 2: The Linked Data Application (LDApp) interface.
Figure 2 presents the LDApp user interface. The
left frame contains the domain graph that is
compiled from ontologies covering three domains:
disease, drug and drug target. The right frame
mainly contains the query results and mechanisms to
assist the user’s search and navigation through them.
In the example depicted in this figure, the term that a
clinician is interested in is the “Myocardial
Infarction” and the domain is a graph based on the
Disease concept of the Diseasome ontology. The
results of DESCRIBE queries are presented to the
users of LDApp at their original form (as pictured in
figure 2); SELECT queries are presented using an
RSS style (Figure 3) with the title, link and
description of each data item.
2.2 Answering Hypothesis Questions
In order to demonstrate the benefits of using the
LDApp, we will use an example that is based on one
of the PONTE use cases. We assume that the
clinician’s question is about clinical trials on the
disease “Myocardial Infarction”. Initially results for
this disease are retrieved from the resource
“Myocardial Infarction” of LD source Diseasome
(see Figure 2).
The Diseasome resource has a list of
diseasome:possibleDrug properties pointing to drug
resources in DrugBank and DailyMed LD sources.
Other links include a list of genes responsible for the
syndrome and owl:sameAs pointers to other data
sources. By selecting one of these properties (RDF
links) the clinician is effectively constructing queries
to Linked Data sources. For example s/he is
interested in possible drugs against the disease
(diseasome:possibleDrugs), so s/he selects
“possibleDrugs” from the expansion list (of figure
2), which triggers a dynamically created query,
which in this example is the following:
SELECT DISTINCT ?drug ?description
WHERE{
?x a diseasome:diseases.
?x diseasome:name ?name.
?x diseasome:possibleDrug ?drug.
OPTIONAL {?drug
drugbank:description ?description.}
FILTER regex (?name,"Myocardial",i)}
Figure 3 shows how the results of this query are
presented in the LDApp. For example, the drug
Etanercept is among the results of the above query
along with data linked to it such as its description,
indications, other diseases for which it can be used
as treatment, clinical trials, side effects and much
more. This additional information on each of the
resulting items is presented to the clinician on the
right frame of LDApp when s/he clicks on its title of
the item.
Additionally, the left frame presents the
“Etarnecept” instance in the DrugBank ontology
again with some expansion options. The clinician
can continue her/his search and choose an
owl:sameAs expansion; results from several Linked
Data sources are returned that can be of great
interest to the specification of study parameters:
such as from SIDER, a source for side effects and
from ClinicalTrial.gov (LinkedCT Linked Data
source) with descriptions on past clinical trials of
this drug. Hence, this process can facilitate the
clinician’s work in finding/extracting the
information that s/he needs.
3 CONCLUSIONS AND FUTURE
WORK
In this article, we presented a prototype application
that consumes Linked Data for assisting the design
process of drug repositioning clinical trials. We have
explained our approach for aggregating information
from heterogeneous data sources by exploiting the
links between them, which allows navigation and
ALinkedDataApproachforQueryingHeterogeneousSources-AssistingResearchersinFindingAnswerstoComplex
ClinicalQuestions
413

Figure 3: Querying Diseasome and exploiting the possibleDrug link.
performing queries across different data sources, and
by assisting users to navigate from one topic (data
source) to another using the ontology based
navigation frame. In order to demonstrate the
capability of answering research questions for the
test of hypothesis and study parameters specification
we have developed a scenario in which the Linked
Data Application (LDApp) provides initial results
and then extends clinical researcher’s search
requests using the Web of Linked Data.
We have plans to extend the use of Linked Data
Application and Query Engine to other processes of
the PONTE platform such as the validation of the
clinical trial protocol and the proposing of eligibility
criteria. We are also looking at further improving the
query engine and especially developing a “query
expansion” mechanism. This mechanism aims at
reformulating a seed query to improve retrieval
performance in information retrieval operations
(Hartig, 2012) and, in the context of semantic data, it
will involve performing additional searches in
semantically associated data sources.
REFERENCES
Ashburn, T., Thor, K., 2004. Drug repositioning:
identifying and developing new uses for existing
drugs. Nature reviews. Drug discovery.
Belleau, F., Nolin, M. A., Tourigny, N., Rigault, P.,
Morissette, J., 2008. Bio2RDF: towards a mashup to
build bioinformatics knowledge systems, Journal of
biomedical informatics, S.706-716,
Berners-Lee, T. 2006. Linked Data - Design Issues. http://
www.w3.org/DesignIssues/LinkedData.html
Bizer C., T., Heath, T., Berners-Lee, T., 2010. Linked
data-the story so far, sbc, S.9
Bouquet, P., Ghidini, C., Serafini, L., 2009. Querying the
Web of Data: A formal approach. In Proc of the 4th
Asian Semantic Web Conference (ASWC).
Chondrogiannis, E., Matskanis, N., Roumier, J., Massonet,
P., Andronikou, V., 2011, Enabling semantic
interlinking of medical data sources and EHRs for
clinical research purposes, eChallenges conference
Hartig, O., Bizer, C, Freytag, J. C., 2009. Executing
SPARQL Queries over the Web of Linked Data, In
Proceedings of the 8th International Semantic Web
Conference (ISWC), Washington, DC, USA.
Hartig, O., Freytag, J. C., 2012: Foundations of Traversal
Based Query Execution over Linked Data. In
Proceedings of the 23rd ACM Conference on
Hypertext and Social Media (HT), Semantic Data
Track, Milwaukee, WI, USA
PONTE Project, Efficient Patient Recruitment for
Innovative Clinical Trials of Existing Drugs to other
Indications, http://www.ponte-project.eu/
Samwald, M., Jentzsch, A., Bouton, C., Kallesøe, C. S.,
Willighagen, E., Hajagos, J., Marshall, M. S.,
Prud'hommeaux, E., Hassanzadeh, O., Pichler, E.,
Stephens, S., 2011. Linked open drug data for
pharmaceutical research and development, Journal of
Cheminformatics, 3:19 doi:10.1186/1758-2946-3-19
Schmidt, M., et al., 2011. FedBench: A Benchmark Suite
for Federated Semantic Data Query Processing, ISWC
KEOD2012-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
414
