
BRIDGING THE GAP BETWEEN CLINICAL RESEARCH
AND CARE
Approaches to Semantic Interoperability, Security & Privacy
Richard Vdovjak
1
, Brecht Claerhout
2
and Anca Bucur
1
1
Philips Research, High Tech Campus 34, 5656AE, Eindhoven, The Netherlands
2
Custodix, Kortrijksesteenweg 214 b3, 9830, Sint-Martens-Latem, Belgium
Keywords: Clinical trials, EHR, Semantic interoperability, Security, Privacy, Information integration.
Abstract: Efficient collaboration and data sharing are essential prerequisites for improving efficiency, safety and
outcomes in medicine. Current separation of clinical research and care creates a significant knowledge gap,
especially in the case of complex diseases such as cancer, hampering research and slowing down the
transfer of the latest research results to patient care. The momentum gained by initiatives focusing on these
aspects indicates that under the right circumstances, the biomedical community is ready and willing to open
up. However, main technological barriers concerning semantic interoperability, security and privacy need to
be addressed to make this change possible. In this paper we describe our scalable, standards-based and open
approach towards addressing these issues in the context of a large initiative with focus in oncology.
1 INTRODUCTION
Despite large investments in IT, the healthcare
domain is currently unable to obtain the desired
benefits in quality, safety and efficiency of care and
to use those IT systems at their full potential. The
lack of integration and of semantic interoperability
among systems is a significant source of
inefficiency, data inconsistencies, unnecessary costs
and an unacceptably large number of medical errors.
As the cost of healthcare in Europe becomes almost
unaffordable, reducing expenses while significantly
increasing the quality, safety and efficiency of care
is a necessity. Furthermore, the pharmaceutical
industry faces low recruitment rates of patients and
extremely high costs of running clinical trials due to
lack of interoperability and complex and inefficient
study execution, while having a strong need to
reduce research expenses and the time-to-market of
new drugs.
Additionally, there is a widening knowledge gap
between the care provided in top research clinical
sites and standard care sites, resulting in large
differences in treatments and outcomes. In this
context, the need to bring the latest therapy options
validated in clinical research to each and every
hospital must be addressed before being able to
significantly reduce the numbers of patients that
receive suboptimal treatment (e.g. overtreatment,
wrong dose, etc.), or the wrong treatment. There are
currently very few mechanisms and formally
established channels for transferring the best
practices to clinicians and the current dissemination
means are insufficient.
While the need to share and collaborate is
increasingly being recognized, with large initiatives
gaining significant support
1
,
2
, several technological
issues limit progress: lack of semantic
interoperability among systems in care and research,
and concerns regarding security and privacy if those
systems were to open up.
In this paper we describe our approach towards
semantic interoperability, which will be
implemented part of a large EU-funded initiative and
deployed within a broad community of top European
healthcare organizations that focus on research and
care in oncology. We aim to enable seamless,
secure, scalable and consistent linkage of healthcare
information residing in EHR systems with
information in clinical research information systems,
such as clinical trial systems, supporting the two
1
www.ecrin.org
2
www.breastinternationalgroup.org/
281
Vdovjak R., Claerhout B. and Bucur A..
BRIDGING THE GAP BETWEEN CLINICAL RESEARCH AND CARE - Approaches to Semantic Interoperability, Security & Privacy.
DOI: 10.5220/0003872202810286
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2012), pages 281-286
ISBN: 978-989-8425-88-1
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

currently separated worlds of clinical research and
clinical practice to connect and benefit from each
other.
The remaining of the paper is structured as
follows. Section 2 argues for the need to provide the
appropriate technological solutions to enable
semantic bi-directional linkage of clinical care and
clinical research data. We also describe relevant
applications in research and care that would benefit
from a scalable and secure semantic interoperability
solution. Our approach to semantic interoperability
is described in Section 3. We also address the
privacy and security needs related to sharing patient
data across research and care, as described in
Section 4.
Figure 1: From implementation heterogeneity to semantic
agreement.
2 NEED FOR LINKING EHR AND
CLINICAL TRIAL DATA
The need for integration of clinical care and clinical
trial systems has been identified as a way to
significantly improve the effectiveness and
efficiency of clinical research (Ohmann, 2007). We
believe that such integration can also strongly
benefit clinical care. Next to benefits concerning
patient outcomes and safety, this integration has an
important potential to bring along significant cost
reduction.
The current separation between clinical research
and clinical care makes the detection of many
serious patient safety issues difficult. Serious side
effects (Oeffinger, 2009), of therapy and drugs that
appear outside a clinical trial either due to a low
incidence or a late onset are very difficult to detect
and to explain in the absence of a feedback loop
from standard care to research.
Although having the potential to bring important
benefits (Safran, 2007), (Pakhomov, 2007), the
secondary use of care data for research, quality
assurance and patient safety is still rarely supported.
Main barriers to enabling secondary use of data are
the lack of interoperability, common standards and
terminologies, and challenges around data security
and patient privacy.
The semantic bi-directional linkage of clinical
research and clinical care systems will support many
highly relevant applications in research and care,
such as:
Supporting more effective and efficient
execution of clinical research by allowing faster
eligible patient identification and enrolment in
clinical trials, and providing access – in a legally
compliant and secure manner – to the large amounts
of patient data collected in the EHR systems to be
re-used in clinical research, for new hypotheses
building and testing (e.g. to benefit rare diseases),
study feasibility, as well as for epidemiology studies.
Enabling long term follow up of patients, beyond
the end of a clinical trial.
Avoiding the current need for multiple data entry
in the various clinical care and research systems
during the execution of a study.
Allowing data mining of longitudinal EHR data
for early detection of patient safety issues related to
therapies and drugs that would not become manifest
in a clinical trial either due to limited sample size or
to limited trial duration, and eliminate duplicate
reporting (in care and research) of identified serious
side effects,
Supporting faster transfer of new research
findings and guidelines to the clinical setting (from
bench-to-bedside).
Figure 2: The core dataset covering a chosen clinical
domain.
Figure 1 depicts our gradual approach towards
reaching semantic agreement among EHR and CT
HEALTHINF 2012 - International Conference on Health Informatics
282

systems. We move away from the implementation
heterogeneity of the local sources by building
canonical models of the sources which will be
mapped to our reference model and to the semantic
core dataset (described in section 3.1). The
canonical models describe the data in the sources
while making use of the semantics of the core
dataset. Figure 2 presents the envisioned application
services enabled by the desired linkage between the
EHR and CT systems.
Figure 3: An overview of enabled application services.
3 SEMANTIC
INTEROPERABILITY
Using the SemanticHEALTH classification
(Stroetmann, 2009) of semantic interoperability
(SIOp) we can observe that the current level of SIOp
between CTs and EHRs is somewhere between level
0, i.e. no interoperability at all, and level 1 i.e.
syntactic interoperability. The reason for this is the
fact that these systems were designed as information
silos in isolation, not foreseeing the benefits of
mutual data exchange as laid out in the section
above. In order to achieve the aforementioned
benefits, we have to increase the SIOp level to at
least 2b - bidirectional semantic interoperability of
meaningful fragments, or even level 3 which
requires full semantic interoperability, sharable
context. It is however also recognized that due to the
steep investments needed, the highest level of
semantic interoperability should only be sought in
specific areas with high potential for significant
improvements.
The essential steps for achieving this SIOp
improvement include the definition of sound
information models describing the clinical trial
systems, building on existing research results when
possible (Weiler
,
2007). Electronic health records
too need to be properly modelled; to that end we will
adopt the appropriate state-of-the-art representation
formalisms such as HL7 CDA, the openEHR
Reference Model, ISO/EN 13606, etc.
3.1 Semantic Core Dataset
The foundation of the semantic interoperability layer
will be the semantic core dataset comprising soundly
defined and agreed-upon clinical structures
consisting of standard-based concepts, their
relationships, and quantification (e.g. archetypes
using selected terminology concepts) that together
sufficiently describe the semantics of the chosen
clinical domain.
The semantics of the clinical terms should be
captured by standard terminology systems such as
SNOMED CT, ICD, LOINC. The scalability of the
solution needs to be achieved by modularization and
scoping, e.g. instead of aiming at inclusion of the
complete SNOMED terminology (more than 300
thousand concepts) we identify a core subset that
covers the chosen clinical domain. The main
rationale here is that only a confined subset of
relevant concepts from the clinical ontology will be
needed for data extraction and reasoning in a given
clinical context/domain while most of the remaining
concepts would never be used by reasoning
algorithms.
Such core dataset shall be validated both by
clinical and knowledge engineering experts to assure
proper coverage and soundness. In the process of
identifying the core data set and the corresponding
mapping tools, care will be taken to allow for easy
extension of the core data set, should the inclusion
of new concepts become necessary (e.g. a cross-
domain linkage). Relying on well established and
widely used existing terminology standards will
facilitate extensible semantic interoperability
towards third parties outside of the scope of the
project. This approach is in line with the roadmap of
SemanticHEALTH which lists identifying of sound
semantic subsets of SNOMED covering a certain
clinical domain as one of their priorities
(Stroetmann, 2009).
The core semantic data set will be validated in
concrete use cases, for the different EHR and
clinical trial systems available at the clinical care
and clinical trial sites within the consortium. The
semantic core dataset is an essential prerequisite to
semantically-aware access to both EHR and Clinical
trial data in a machine processable manner.
Concepts in the dataset will have their unique
identifiers, well understood meaning, as well as a set
of synonyms they can be referred as.
BRIDGING THE GAP BETWEEN CLINICAL RESEARCH AND CARE - Approaches to Semantic Interoperability,
Security & Privacy
283
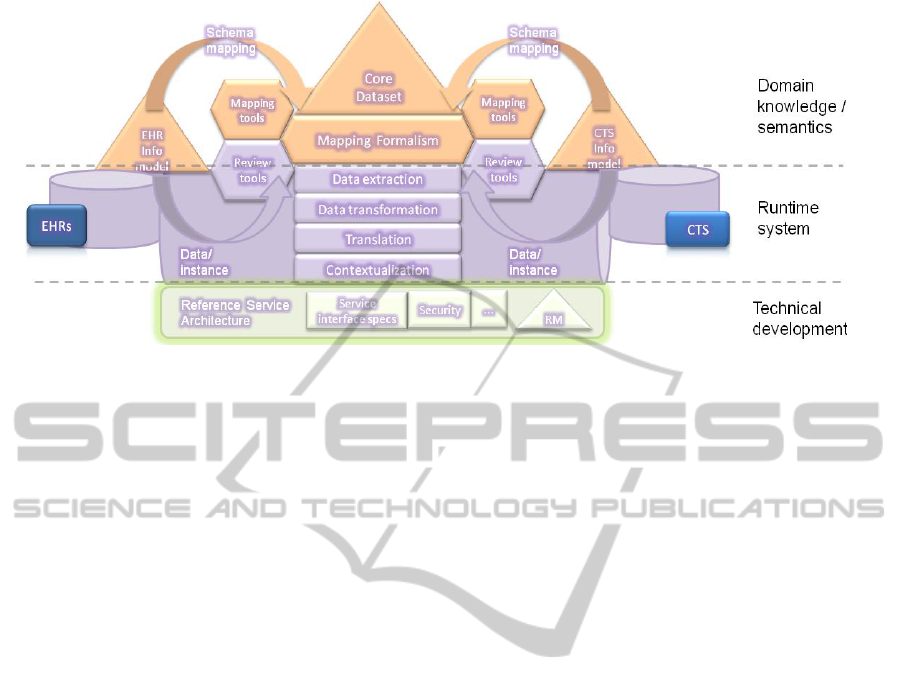
Figure 4: Schema-level mapping definitions (top), the underlying semantic interoperability run-time platform that handles
the necessary data transformations (middle), and the underpinning technical blueprint including the Reference Architecture
and the Reference Model (bottom).
Multicenter clinical trials often span across
several countries which introduces the problem of
language heterogeneity between the CTs and EHRs
as primary data capture. We plan to address this
issue by offering a gradual approach, semi-
automatically translating only those parts of the
clinical ontology identified as the core semantic
dataset, leveraging existing translations of known
terminologies such as SNOMED CT. When no
translation of the relevant standard terminologies
exist in that language, we will work out together
with the clinical experts a translation of the core
dataset into the languages that are used for the
primary data capture. Hence, translating (only) the
selected semantic core dataset and not the entire
clinical coding system enables a modular and
scalable approach where the initial translation effort
is limited in scope and delivers immediate benefits
in increased semantic interoperability.
3.2 The Semantic Interoperability
Platform
The canonical information models of the EHR and
CT systems will be mapped to the semantic core
dataset in order to guarantee a well defined meaning
of various data elements across the entire platform.
We will identify the requirements for mappings that
bridge the semantic core data set with the
information models representing the EHR systems
and the clinical trial systems. These information
models provide a canonical view, reflecting the
content and the structure of the respective
information management system. The proposed
mapping formalism should be able to mitigate the
foreseen structural and contextual differences
between the semantic core dataset and the
information models. We will use this formalism to
instantiate the necessary schema-level mappings
(Figure 4, top) that will be executed by the semantic
interoperability platform during the data extraction
process.
In order to facilitate the necessary data
transformation among various information systems
which need to interoperate, we deploy our semantic
interoperability run-time platform (Figure 4,
middle). This platform will utilize the semantic core
dataset as well as the schema-level mappings that
link to the EHR and CT information models. The
platform will be able to execute these mappings
during the data extraction phase, instantiating thus
the semantic concepts with patient data and/or
clinical trial data. The semantic interoperability
platform will be an essential software engine behind
the application services, enabling linkage between
the patient data in the EHR and the clinical trial
systems. The platform itself is an instantiation of our
reference service architecture blueprint and
leverages the chosen reference model (RM) (Figure
4, bottom).
4 SECURITY AND PRIVACY
The sensitive nature of health information and the
harm that can be caused by its abuse is widely
known. It needs no debate that the risk and impact of
this abuse significantly increases when more
information about individual patients is accumulated
and is more frequently exchanged among different
HEALTHINF 2012 - International Conference on Health Informatics
284

parties (caregivers, researchers, etc.). Specific
legislation, regulations and ethical guidelines with
respect to (patient) privacy have therefore been put
in place at different levels (European, national and
regional).
In this context, the capability to satisfy varying
ethical concerns and ensure compliance to data
protection legislation and regulations is fundamental
to the success (viability in the long run) of any
solution aiming to integrate health information on a
large scale.
Our approach to this matter comprises the design
of a comprehensive Data Protection Framework
(DPF) which outlines the boundaries within which
services (and organisations) are required to operate.
The DPF brings “compliance by design” by
combining both a governance framework (policies
and procedures) and a set of technical
implementations aimed at enforcing the latter. It
implements the rules set by the relevant National
and EU legislation and sector best practice policies
(ethics). The framework not only manages and
enforces rules defining “Who has access to what
data for which purpose, and under what conditions”,
but also integrates solutions which enable access to
otherwise unavailable data (a.o. Trusted Third Party
supported de-identification).
Introducing a uniform layer (technical solutions
integrated in a single governance framework) upon
which applications can (and need to) build has
already proven to be a successful approach
(Claerhout, 2008) to efficiently deal with regulatory
issues of large scale transnational sharing of medical
and biological data in the clinical trial context. One
of the things that the overall governance and security
framework referenced above introduced was a novel
practical solution (concept of “de-facto anonymous
data”) that covers the inherent issues tied to de-
identification of individual person records (Li,
2007). That work will serve as a basis for our DPF
which needs to deal with the broader scope of bi-
directional cross-domain interaction between the
care and research domain.
Technically, the DPF will rely on (centralised)
policy based authorization services to translate the
legal rule sets into authorisation decisions for
“access to” or “processing of” highly sensitive data
over distributed resources. This approach ensures
flexibility towards changing legislation and policies
(and regional variations thereof).
To meet the specific requirements of the DPF,
the authorisation system (both decision and
enforcement parts) needs to support concepts such as
“purpose of use” and “conditions on use” (e.g. by
introducing sticky policies (Chadwick, 2008)
associated with datasets, or other types of privacy-
metadata) and work at least at the granular level of
“a logical dataset”. Meeting these requirements in a
generic (loosely coupled) way and with sufficient
performance is challenging.
Patient consent is another important aspect which
is unmistakably connected to data protection, for
example with respect to re-use of personal data
beyond its originally intended use (e.g. use of EHR
data for automated eligibility scanning, for export
for research purposes, etc.). Technically, “Consent
Management Services” fit into the framework as
specialised authorization services (consent rules
form a policy). Such services need to ensure the
integrity of consent directives and correctly combine
them to avoid conflicting preferences.
Complementary to preventive security measures,
the framework requires audit mechanisms allowing
detection of security breaches and data leakage (and
tools for subsequent incident handling).
Currently, the majority of auditing mechanisms
log individual events per application or computer
system. In order to reconstruct a logical chain of
events for proper audit in large distributed networks,
these different logs would need to be combined. Few
standards and solutions are available providing
manageable uniform audit trails in distributed
systems.
Furthermore, to be useful for checking
compliance of a (large) system with data protection
legislation, audit trails need to include extended
contextual information, which they rarely do (e.g.
type of data accessed, identity of the person listed in
the medical record accessed, etc.). Moreover, logs
need to be readily accessible in a user-centric and
data-centric way (e.g. be able to give an overview of
activity of a single user throughout the network or
the actions performed on a specific logic dataset).
Reconstruction of such user-centric or data-centric
audit trails based on standard logs is typically not
feasible in practice: audit trail data is too large to
efficiently query, identity of data subjects is not
recorded or cannot be linked across applications, etc.
In order to undeniably assess the compliance of
data flows with regulations, the provenance of
received information and stored data must be
recorded. Knowing the provenance of a data set can
for example inform a user or system about the
applicable data privacy policies (cf. consent). But
provenance goes beyond security, and for one plays
a very important role in data quality management
(who is the original source, how was it recorded,
cleansed, transformed, etc.).
BRIDGING THE GAP BETWEEN CLINICAL RESEARCH AND CARE - Approaches to Semantic Interoperability,
Security & Privacy
285

Extended audit and provenance functionality
thus comprises an important part of the technical
framework.
5 CONCLUSIONS
The momentum gained by new initiatives focused on
data sharing and collaboration
3
,
4
indicates that under
the right circumstances, the biomedical community
is willing to open up. We aim to support this
important culture shift by building the necessary
environment that will provide the needed level of
semantic interoperability in full compliance with
security, privacy and legal requirements.
Interoperability is by definition a global issue
which cannot be successfully tackled in isolation,
requiring both critical mass and openness.
Therefore, to ensure a low barrier to adoption within
a large community, we adopt a pragmatic approach:
We rely on collaborative effort and propose a
modular development of the semantic core dataset,
which makes use of ontologies (e.g. SNOMED CT)
and standards (e.g. HL7) that benefit of significant
use in healthcare.
Our Data Protection Framework aims to provide
a “unified” solution for achieving regulatory
compliance (privacy & security) “by design” with
minimal effort to anyone subscribing to the proposed
integrating architecture.
REFERENCES
Ohmann, C., Kuchinke, W, 2007. Meeting the Challenges
of Patient Recruitment. A Role for Electronic Health
Records. In Int. J. Pharm. Med.
Oeffinger, K.C., 2009. Breast Cancer Surveillance
Practices Among Women Previously Treated With
Chest Radiation. In JAMA 301: 404-414.
Pakhomov, S., 2007. Electronic medical records for
clinical research: application to the identification of
heart failure. In Am J Managed Care.
Safran, C., 2007. Toward a national framework for the
secondary use of health data: an American Medical
Informatics Association white paper. In JAMIA.
Stroetmann, V., Kalra, D., Lewalle, P., Rector, A.,
Rodrigues, J., Stroetmann, K., Surjan, G., Ustun, B.,
Virtanen, M., Zanstra, P., 2009. Semantic
Interoperability for Better Health and Safer
Healthcare, SemanticHEALTH Report, pp. 12-13.
3
https://cabig.nci.nih.gov/nci-ncri2010conference/Esserman.pdf
4
http://sagebase.org/
Weiler, G., Brochhausen, M., Graf, N., Schera, F., Hoppe,
A., Kiefer, S., 2007. Ontology Based Data
Management Systems for post-genomic clinical Trials
within an European Grid Infrastructure for Cancer
Research. In Proc of the 29th Annual Int. Conf. of the
IEEE EMBS.
Claerhout, B., Forgó, N., Krügel, T., Arning, M., De
Moor, G., 2008; A data protection framework for
trans-European genetic research projects, Stud Health
Technol Inform, pp. 67-72.
Li, N., Li, T., Venkatasubramanian, S.: t-closeness:
Privacy beyond k-anonymity and l-diversity. In: Proc.
IEEE Int. Conf. Data Eng. (ICDE), Istanbul, Turkey
(April 2007) page 106-115.
Chadwick, D., Lievens, S., Enforcing "Sticky" Security
Policies throughout a Distributed Application.
http://www.cs.kuleuven.be/conference/MidSec2008/sti
cky.pdf.
HEALTHINF 2012 - International Conference on Health Informatics
286
