
MELANOSOME TRACKING BY BAYES THEOREM
AND ESTIMATION OF MOVABLE REGION
Toshiaki Okabe
and Kazuhiro Hotta
Meijo University, 1-501 shiogamaguchi, Tenpaku-ku, Nagoyasi, 468-8501, Japan
Keywords: Tracking, Intracellular image, Melanosome, Systems biology.
Abstract: This paper proposes a melanosome tracking method using Bayes theorem and estimation of movable region
of melanosome candidates. Melanosomes in intracellular images are tracked manually now to investigate
the cause of disease, and automatic tracking method is desired. Since there are little automatic recognition
methods for intracellular images, we can not know which features and classifiers are effective for them.
Thus, we try to develop the melanosome tracking using Bayes theorem of melanosome candidates detected
by Scale-Invariant Feature Transform (SIFT). However, SIFT can not detect the center of melanosome
because melanosome is too small in images. Therefore, SIFT detector is adopted after image size is enlarged
by Lanczos resampling. However, there are still many melanosome candidates. Thus, we estimate the
movable region of the target melanosome in next frame and eliminate melanosome candidates. After the
posterior probability of each candidate is computed by Bayes theorem, and the melanosome with the
maximum probability is tracked. Experimental results using the melanosome images of normal and Griscelli
syndrome show the effectiveness of our method.
1 INTRODUCTION
Live cell imaging is advanced rapidly in recent years
because of the progress of microscope techniques
(Sakaushi et al., 2007; Sugimoto and Tone, 2010;
Sakaushi, et al., 2008). Especially, elucidation of
transportation path in cells is very important for
understanding of clinical state. However, there are
little automatic recognition methods for live cell
imaging, and human counts and tracks the particles
in cells manually now. This work is hard physically
and mentally for humans, and human can not treat a
lot of data. Since many objective data are required
for the investigation into the cause of disease,
automatic recognition methods for intracellular
images are desired. Therefore, we try to develop a
tracking method for intracellular images. This is new
application of computer vision and very contributes
to medical development.
In this paper, the tracking target is the
melanosome in the melanocytes (Kuroda, et al.,
2003) (Kuroda, et al., 2004). The melanocyte
combines the melanin pigment and stores in cell
membrane which is called melanosome . The
melanosome is transported into cells. It is known
that transport disorder causes abnormal pigmentation.
Thus, the melanosome tracking is important for the
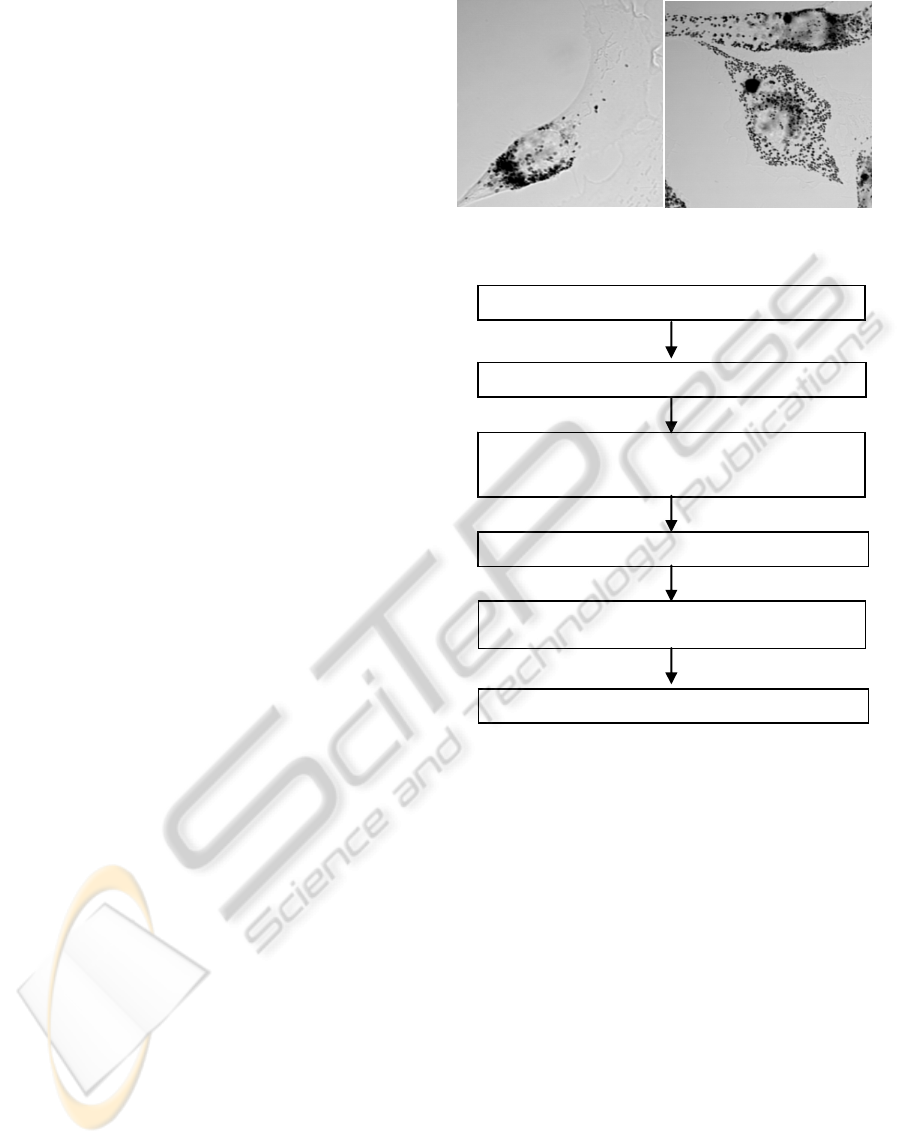
investigation into the cause of disease. Figure 1
shows the examples of melanosome images in which
melanosome is the particle with black color. We
must track a particle which is not different from
neighboring particles. If we can realize an automatic
melanosome tracking method, it will be applicable
to particle tracking in various kinds of cells.
There is a software which is usually used in cell
biology. That is called SpotTracker2D which is
plugin of imageJ. We try to track melanosome by
using the SpotTracker2D but it can not track
melanosome well. There are not any conventional
studies about automatic melanosome tracking by
computer. we can not know which features and
classifiers and effective for melanosome tracking.
Since we do not have any clues, we divide the
melanosome tracking into 2 tasks. The first task is
the candidate detection. The second task is the
posterior probability estimation of the candidates.
In the first task, we try to detect the melanosome
candidates by SIFT (Lowe, 2004). However, SIFT
can not detect the center of melanosome because
melanosome is too small in the original microscope
images. Therefore, SIFT is adopted after original
482
Okabe T. and Hotta K. (2012).
MELANOSOME TRACKING BY BAYES THEOREM AND ESTIMATION OF MOVABLE REGION.
In Proceedings of the 1st International Conference on Pattern Recognition Applications and Methods, pages 482-487
DOI: 10.5220/0003836104820487
Copyright
c
SciTePress
microscope images are enlarged by Lanczos
resampling (Duchon, 1979) (Turkowski and Gabriel,
1990). However, SIFT also detects the characteristic
points on non-melanosome by image enlargement.
This may induce tracking failures. Thus, we must
eliminate the casdidates. Thus, the movable region
of a tracking target in the next frame is estimated
under the assumption that the target melanosome
does not pass through other melanosomes.
In the second task, the posterior probability of
every candidate which is selected by upper steps is
computed by Bayes theorem, and the candidate with
maximum probability is tracked. However, SIFT
detects some features on the same position.
Therefore, the posterior probabilities of all features
on the same position are computed independently,
and the posterior probabilities on the same position
are integrated by sum or product.
The melanosome images of normal and Griscelli
syndrome are used in experiments. The accuracy
achieves 94.4% when the position of melanosome at
time t+1 is predicted from that at time t. The image
enlargement by Lanczos resampling, the estimation
of movable region and sum of the posterior
probability are effective for this task.Although the
accuracy decreases in the task that the correct
position of melanosome in only the first frame is
given and the positions in the remaining frames are
predicted, the possibility of our method is
demonstrated by experiments.
This paper is constructed as follows. In section 2,
the details of the proposed method are explained.
Experimental results are shown in section 3. Finally,
conclusions and future works are described in
section 4.
2 PROPOSED METHOD
Figure 2 shows the flowchart of the proposed
method. Candidates detection and feature of
candidates are required for melanosome tracking.
However, recognition techniques for intracellular
images are not established and conventional
methods do not exist! Thus, in this paper, we use
characteristic feature point detection and descriptor
by SIFT. However, feature points are not detected at
the center of melanosome because the melanosome
is too small in the original microscope images.
Therefore, the image size is enlarged by Lanczos
resampling, and SIFT is applied to the image
enlarged 9 times (Okabe and Hotta, 2011). However,
SIFT also detects the feature points on non-
melanosomes. Thus,
Figure 1: (a) Intracellular image of normal melanocyte (b)
Intracellular image of Griscelli syndrome melanocyte.
Figure 2: Flowchart proposed method.
melanosome candidates are eliminated by
binarization of intensity because the color of
melanosome is black. In addition, we estimate the
movable region of target melanosome in the next
frame under the assumption that the target
melanosome does not pass through other
melanosomes. The posterior probability of
remaining candidates are computed by Bayes
theorem, and the position
with maximum
posterior probability is tracked (Okabe and Hotta,
2010). Each element in the proposed method is
explained in the following sections.
2.1 Scale-Invariant Feature Transform
SIFT is an algorithm for detecting characteristic
feature points and for describing the detect points.
The characteristic feature points are robust to
rotation, scaling and brightness change. Melanosome
candidates are detected by SIFT, and SIFT
Image enlargement by Lanczos resampling
Posterior probability by Bayes theorem
Location estimation of melanosome
Candidates elimination by intensity binarization
Melanosome candidate detection by SIFT
Estimation of movable region
(a)
(
b
)
MELANOSOME TRACKING BY BAYES THEOREM AND ESTIMATION OF MOVABLE REGION
483

descriptor is used as the feature for computing the
probability.
2.2 Image Enlargement by Lanczos
Resampling
Since, melanosomes in images are very small, SIFT
can not detect correct position. Therefore, image
enlargement by Lanczos resampling used to detect
correct position of melanosomes.
Lanczos kernel is constructed by the product of 2
sinc functions.
L
x
sinc
x
sinc
x
a
ax,0
1 x0
0 otherwise
(1)
Sinc function is defined as
π
π
. The value of "a"
is usually set to 2 or 3. When "a" is bigger than the
absolute value, L
x
0. When x=0, L(x)=1 [15].
Equation (1) can be redefined as
sinc
x
sinc
x
a
asinπxsin
πx
a
π
x
.
(2)
The 2 dimensional Lanczos kernel can be made by
the product of 1 dimensional kernel. The enlarged
image I
x
,y
is computed as
I
x
,
y
I
i,j
L
x
i
L
y
j
.
(3)
Since Lanczos kernel of a=3 gives clear image
enlargement, we use a = 3 in experiments.
2.3 Candidates Elimination by
Intensity Binarization
SIFT can detect the center of melanosome by image
enlargement. However, SIFT also detects non-
melanosome regions as shown in Figure 3 (c). Since
the color of melanosome is black, non- melanosome
candidates with white color are eliminated by
intensity binarization. The elimination of candidates
will improve tracking accuracy.
When threshold is θ, binarization result g
x,y
is
defined as
g
x,
y
=
1
f
x,
y
θ
0
f
x,
y
θ
.
(4)
This threshold value affects tracking accuracy.
Therefore, we determine the threshold value
experimentally. We evaluate the tracking accuracy
for 12 melanosomes for parameter estimation by
change the threshold value. Note that these 12
Figure 3: (a) Intracellular image (b) Result of intensity
binarization (c) Feature poins detected by SIFT are shown
as diamond shape (d) SIFT feature poins after intensity
binarization.
melanosome are not used in test. We found that
θ = 100 gives the best accuracy, and θ is set to 100
in the following experiments. Figure 3 (b) is the
example of binarization result. Figure 3 (c) is the
result of SIFT detector in terms of the image before
candidates elimination. On the other hand, Figure 3
(d) shows the result of SIFT detector after
candidates elimination. We understand that non-
melanosome candidates with white color are
eliminated in Figure 3 (d).
2.4 Location Prediction by Bayes
Theorem
Next we explain there to compute the posterior
probability of each candidates. SIFT descriptor with
128 dimensions obtained at the characteristic point
is used as the feature
for computing the
posterior probability of
. Since we treat the
tracking problem, the posterior probability in terms
of
,…,
is considered. Posterior probability
P
|
is computed as
P
|
p
|
,
p
|
p
|
.
(5)
We assume that
is independent of
,…,
.
Thus, p
|
,
can be written as p
|
,
p
|
can be written by using the posterior
probability at time t-1 and transition probability as
p
|
p
|
p
|
d
.
(6)
Then, the posterior probability P
|
can be
(c)
(a)
(
b
)
(
d
)
ICPRAM 2012 - International Conference on Pattern Recognition Applications and Methods
484

written as
P
|
p
|
,
p
|
p
|
d
p
|
.
(7)
We compute the posterior probability of all
candidates and track the location with maximum
probability. We do not need to compute p
|
because it is normalization coefficient.
Conditional probability and transition probability
are explained in the following sections.
2.4.1 Conditional Probability
The features of the same melanosome between time
t and t-1 are not so changed. Thus, conditional
probability is modelled as normal distribution using
the SIFT features at time t and t-1 as
p
|
,
1
√
2πσ
exp
‖
‖
2σ
,
(8)
Where
is 128 dimensional SIFT descriptor on
location
at time t and
is SIFT descriptor at
time t-1.
2.4.2 Transition Probability
The location of tracking target at time t and t-1 is
denoted as
w
,h
and
= (w
,h
). If
we assume that the melanosome does not move so
far from time t-1 to t, transition probability is also
modelled by the normal distribution as
p
|
1
√
2πσ
exp
w
w
h
h
2σ
,
(9)
Equation (9) limits the movable region because
transition probability becomes small for far points
from
.
2.5 Estimation of Movable Region
Figure 3(a) shows that there are many melanosomes
with similar appearance in a local region. This
Figure 4: (a) Intracellular image after intensity
binarization (b) Movable region of tracking target shown
as the red circle in Figure 4(a).
induces tracking failure. The similarity measure by
SIFT may not be sufficient. Thus, we estimate the
movable region of a tracking melanosome in next
frame to improve the accuracy. If we assume that
other melanosomes except for the tracking target do
not move, we can estimate the movable region of the
target. First, we binarize the image as shown in
Figure 4(a). Melanosome is represented as black
color and the non-melanosome region is represented
as white color. The target melanosome is shown as
red circle. If target melanosome does not pass
through other melanosomes and other melanosomes
do not move, we can estimate the movable region as
shown in Figure 4(b). The white regions are the
movable region in the next frame. Experimental
results show that the estimation of movable region
decreases the tracking failure.
3 EXPERIMENTS
We use the melanosome images obtained from
Technical Committee on Industrial Application of
Image Processing (http://www.tc-iaip.org/algorithm.
html). The correct positions of melanosomes in these
images are also included. The 44 melanosomes (31
normal and 13 Griscelli syndrome) are used in
evaluation. Note that these melanosomes are
different from the melanosome used for parameter
selection. We evaluate our method in 2 kinds of
tasks. The first task evaluates whether the true
position of melanosome at time t is estimated from
the supervised position at time t-1. The second task
evaluates whether the position of melanosome at
time t is estimated when the supervised position in
only the first frame is given. In the second task, we
use the simplest case of our method in which only
the posterior probability of tracked position at time
t-1 is 1 and that of other positions is 0 because the
computational cost and memory requirement are
large when the posterior probabilities of all
candidates are saved. This corresponds to the case
that the method used in the first task is adopted
continuously without the supervised position in the
previous frame.
In this experiment, we our method is compared
with SpotTracker2D which is usually used in cell
biology and our proposed method without estimation
movable region. SpotTracker2D is a robust tracker
for microscope images (Sage et al., 2005). In
SpotTracker, LoG filter is used to enhance the target
and to reduce noises. After that, target particle is
tracked by using dynamic programming. Table 1
shows the accuracy in the first task. Tracking
(a)
(b)
MELANOSOME TRACKING BY BAYES THEOREM AND ESTIMATION OF MOVABLE REGION
485
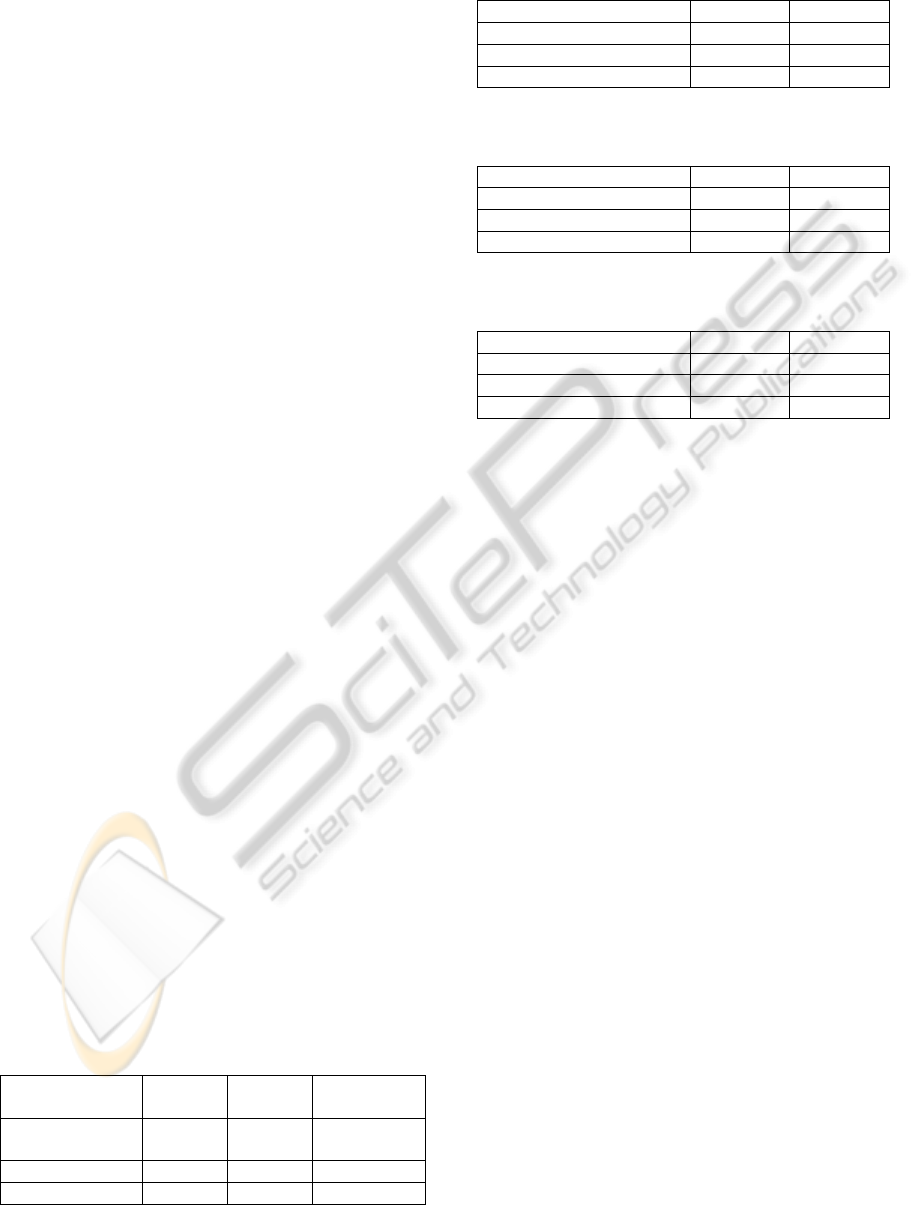
accuracy of our method is much better than
SpotTracker2D. We found that conventional
SpotTracker2D is not useful for the melanosome
tracking and our method using Bayes theorem and
SIFT is effective. Table 2 shows the result of our
method with estimation of movable region. Tracking
accuracy is improved by estimation of movable
region. This shows the effectiveness of the
estimation of movable region. Tables also show that
sum of probability is better than product. It is known
that the integration by sum is effective for several
tasks (Kittler et al., 1998) (Hotta, 2009). The best
accuracy our method achieves 94.4%. Though
SpotTracker2D achieves 73.7%.
In the second task, we do not evaluate
SpotTracker2D because it was much lower than the
proposed method in the first task. Table 3 and 4
show the result of the second task of our method.
Table 3 and 4 show that tracking accuracy for
Griscelli syndrome decreases by using our movable
region. This is because we assume that another
melanosomes except for the tracking target do not
move and melanosomes in Griscelli syndrome move
actively. Thus, tracking accuracy of Griscelli
syndrome decreased slightly. However, the tracking
accuracy of Normal melanosome with estimation of
movable region is better than that without estimation
of movable region. The average of tracking accuracy
is improved.
The accuracy in the second task decreased in
comparison with the first task. This is because one
tracking failure induces the failures in the following
frames. We can consider two reasons for inducing
one tracking failure. The first reason is SIFT
detector. There were some cases that SIFT failed to
detect the target melanosome. The proposed method
can not track the target when the target melanosome
is
not detected as the melanosome candidates. In the
first task, this failure decreases the accuracy slightly.
However, this failure decreases accuracy much more
in the second task because the proposed method
does not have the obvious function for recovering
from the error.
The second reason is the candidate elimination by
intensity binarization. In this paper, threshold value
is determined as 100 which gives maximum tracking
Table 1: Result in the first task without estimation of
movable region.
Product Sum
Spot
Tracker2D
Griscelli
syndrome
91.4% 91.7% 54.6%
Normal 94.6% 94.6% 81.7%
Total average 93.7% 93.8% 73.7%
Table 2: Result in the first task with estimation of movable
region.
Product Sum
Griscelli syndrome 92.2 92.9
Normal 95.0 95.0
Total average 94.2 94.4
Table 3: Result in the second task without estimation of
movable region.
Product Sum
Griscelli syndrome 68.2 70.7
Normal 73.4 73.4
Total average 71.9 72.6
Table 4: Result in the second task with estimation of
movable region.
Product Sum
Griscelli syndrome 66.3 66.5
Normal 78.4 78.4
Total average 74.8 74.9
accuracy in terms of the image set for parameter
estimation. However, this threshold may not be
appropriate for the test set. Some correct
melanosomes were eliminated by intensity
binarization. Thus, one failure induced by
elimination of candidate decreases the accuracy in
the second task. The addition of recovering function
from one failure is the future subject. Although the
accuracy decreases in the second task, the accuracy
achieves 74.9% from only correct position in the
first frame. As you understand from Figure 1, our
there are many similar objects in local region, and
the melanosome is not easy task. The accuracy
demonstrates the effectiveness of the proposed
method.
4 CONCLUSIONS
We proposed a melanosome tracking method using
Bayes theorem and estimation of movable region.
Since SIFT did not work well for the original images,
characteristic feature points are detected after image
enlargement. To improve the accuracy, candidates
are eliminated by intensity binarization and
estimation of movable region. In the first task in
which the position at time t is predicted from the
supervised position at time t-1, the accuracy
achieved 94.9%. This is much better than
SpotTracker2D which is usually used in cell biology.
This shows the effectiveness of our method.
However, in the second task in which melanosome is
tracked in remaining frames from the supervised
ICPRAM 2012 - International Conference on Pattern Recognition Applications and Methods
486

position in only the first frame, one failure induced
the error in remaining frames, and the accuracy
decreased. However, the accuracy achieved 74.9%
for the difficult task in which there are many similar
objects around the tracking target. This demonstrates
the effectiveness of our method for new and
important problem in which conventional methods
are little. This paper will be a giant step for
intracellular image processing.
The future work is to add a recovering function
from tracking failures. We will try the validation
from t to t-1 as well as the position prediction from
t-1 to t.
ACKNOWLEDGEMENTS
We would like to thank Professor Mitunari Fukuda
at Tohoku University and The Japan Society for
Precision Engineering and Technical Committee for
Industrial Application of Image Processing for
providing the opportunity to use melanosome
images. This work was supported by KAKENHI
(23113727).
REFERENCES
T. S. Kuroda, et al., "The actin-binding domain of Slac2-
a/melanophilin is required for melanosome distribution
in melanocytes", Mol. Cell. Biol. 23, pp. 5245-5255,
(2003).
T. S. Kuroda, et al., "Rab27A-binding protein Slp2-a is
required for peripheral melanosome distribution and
elongated cell shape in melanocytes", Nature Cell Biol.
6, pp. 1195-1203, (2004).
D. G. Lowe, "Distinctive image features from scale-
invariant keypoints", International Journal of Computer
Vision, 60, 2, pp. 91-110, (2004).
C. E. Duchon, "Lanczos Filtering in One and Two
Dimensions", Journal of Applied Meteorology 18, 8, pp.
1016–1022, (1979).
K. Turkowski and S. Gabriel, "Filters for Common
Resampling Tasks", In Andrew S. Glassner. Graphics
Gems I. Academic Press, pp. 147–165, (1990).
R. O. Duda, et al., Pattern Classification, JOHN WILEY &
SONS, (2001).
A. I. Dale, Most Honourable Remembrance: The Life and
Work of Thomas Bayes, Springer, (1991).
S. M. Stigler, "Thomas Bayes' Bayesian Inference",
Journal of the Royal Statistical Society, Series A, 145,
pp. 250-258, (1982).
T. Okabe, K. Hotta, "Tracking of Melanosome is Using
Bayes Theorem and SIFT Detector", MIRU 2010
Satellite workshop on Intracellular image processing
pp. 12, (2010). (in Japanese, without review).
T. Okabe, K. Hotta, "Accuracy improvement of
Melanosome by Image Enlargement", IEICE General
Conference, Student poster session, (2011) (in Japanese,
without review).
S. Sakaushi, et al, "Dynamic behavior of FCHO1 revealed
by live-cell imaging microscopy: it's possible
involvement in clathrin-coated vesicleformation",
BiosciBiotechnolBiochem, 71, 7, pp. 1764-1768, (2007).
K. Sugimoto and S. Tone, "Imaging of mitotic cell
division and apoptotic intra-nuclear processes in
multicolor", Methods Mol. Biol. 591, pp. 135-146,
(2010).
S. Sakaushi, et al, "Visualization of aberrant perinuclear
microtubule asterorganization by microtubule-
destabilizing agents", Cell Cycle. 7, 4, pp. 477-483,
(2008).
http://www.tc-iaip.org/algorithm.html.
J. Lund and K. L. Bowers, Sinc Methods for Quadrature
and Differential Equations, SIAM, (1992).
J. Kittler, M. Hatef, R. Dine and J. Matas, "Combing
Classifiers", IEEE Trans. on Pattern Analysis and
Machine Intelligence, 20, 3, pp. 226-239, (1998).
K. Hotta, "Pose Independent Object Classification from
Small Number of Training Samples Based on Kernel
Principal Component Analysis of Local Parts", Image
and Vision Computing, 27, pp. 1240-1251, (2009).
D. Sage, et al, "Automatic Tracking of Individual
Fluorescence Particles: Application to the Study of
Chromosome Dynamics," IEEE Transactions on Image
Processing, 14, 9, pp. 1372-1383, (2005).
MELANOSOME TRACKING BY BAYES THEOREM AND ESTIMATION OF MOVABLE REGION
487
