
SW-ONTOLOGY
A Proposal for Semantic Modeling of a Scientific Workflow Management System
Wander Gaspar, Laryssa Silva, Regina Braga and Fernanda Campos
Computational Modeling Master Program, Federal University of Juiz de Fora, Juiz de Fora, Brazil
Keywords:
Ontology, e-Science, Scientific workflow, Semantic.
Abstract:
The execution of scientific experiments based on computer simulations constitutes an important contribution to
scientific community. In this sense, the implementation of a scientific workflow can be automated by Scientific
Workflow Management Systems, which goal is to provide the orchestration of all processes involved. It aims to
capture the semantic related to the implementation of scientific workflows using ontologies that could capture
the knowledge involved in these processes. Specifically, we present a prototype of an ontology based on
a design pattern called Model View for the representation of knowledge in scientific workflow management
systems.
1 INTRODUCTION
We can consider that scientific research is based on
three pillars: theory, experimentation and computa-
tional resources. The use of these resources helps the
sharing of data, tools and services, and allows the sys-
tematic reuse of experiments. Considering this sce-
nario, researches need an infrastructure that allows the
design, reuse, annotation, validation, sharing and doc-
umentation of the work done by scientists (Barga and
Digiampietri, 2008).
The implementation of experiments based on
computer simulations constitutes an important con-
tribution to the scientific community. In most cases,
current practice is to implement a set of software and
scripts. This procedure has proved insufficient to ad-
equately handle the inherent complexity of the prob-
lems with which scientists have come across. In this
context, it was defined the e-Science term, the sci-
ence that has been largely supported by simulation
and computational infrastructure, based on techniques
like scientific workflows and web services.
In e-Science, a major goal is the creation and use
of processes that simulates experiments, analyzes data
and discovers knowledge, using a wide range of com-
puting resources (Wroe et al., 2007). Technologies
such as ontologies and semantic web services can be
used as the basis for the composition of an infrastruc-
ture to support e-Science (Silva et al., 2009). This pa-
per considers this scenario, presenting the use of on-
tologies and the composition of semantic web servi-
ces in different subdomains in the context of e-
Science. Specifically, we present the SW-Ontology,
an ontology for the knowledge representation in Sci-
entific Workflow Management Systems (SWfMS).
The article is organized as follows. Section 2
presents the background for the research, includ-
ing SWfMS and the use of ontologies for knowl-
edge representation. Section 3 describes some related
works. Section 4 presents the SW-Ontology, a se-
mantic model based on ontologies for scientific work-
flows. Section 5 details the use of SW-Ontology for
the composition of scientific workflows, and finally,
Section 6 presents final considerations and suggests
some future researches.
2 CONCEPTS AND RELATED
WORKS
In a historical perspective, a scientific experiment is
one of the tools used by researchers to support the
formulation of new theories. In this context, a sci-
entific workflow represents the orchestration of pro-
cesses that handle data in order to build a simulation.
2.1 Scientific Workflow Management
Systems
A workflow could be defined as a description of a re-
producible process consisting of a set of interrelated
115
Gaspar W., Silva L., Braga R. and Campos F. (2010).
SW-ONTOLOGY - A Proposal for Semantic Modeling of a Scientific Workflow Management System.
In Proceedings of the 12th International Conference on Enterprise Information Systems - Databases and Information Systems Integration, pages
115-120
DOI: 10.5220/0002865601150120
Copyright
c
SciTePress

tasks (Menager and Lacroix, 2006). The execution of
a scientific workflow can be automated using com-
putational tools called Scientific Workflow Manage-
ment Systems, whose goal is to orchestrate the design,
management and implementation of scientific exper-
iments. The present study intend to capture the se-
mantics involved on the scientific workflows orches-
tration in SWfMS, considering the design process and
the implementation of workflows, using ontologies to
capture the knowledge involved in these processes.
2.2 Ontologies
The word ontology comes from the Greek ontos (be)
+ logos (word). In Philosophy, it is the science of
what is, of the types and structures of objects, proper-
ties, events, processes and relations in every domain.
In this context, the purpose of an ontology is to pro-
vide categorization systems to organize the reality.
Considering Semantic Web, the definition more often
cited in the literature is that an ontology is a formal
and explicit specification of a shared conceptualiza-
tion (Gruber, 1993).
From the 1990s, several languages were proposed
for the representation of ontologies. At the same time,
the rapid expansion of the Internet led to the emer-
gence of lightweight markup languages to support and
at the same time explore the World Wide Web charac-
teristics.
In this context, the Word Wide Web Consor-
tium (W3C) launched and formally recommended as
a standard the Web Ontology language (OWL), de-
signed to meet the requirements of the Semantic Web.
Prot
´
eg
´
e, an editor of ontologies and knowledge bases
supports OWL (Horridge, 2009). In addition, a wide
range of inferences and OWL validation machines are
available, such as Pellet (Sirin et al., 2007) e FaCT++
(Tsarkov and Horrocks, 2006), and semantic Web
frameworks supporting OWL such as Jena (Hewlett-
Packard, 2009).
2.3 Related Work
There are several works related to semantic represen-
tation in scientific workflows. The main contribution
of our work is the use of SW-Ontology in shaping the
composition of scientific workflows in the context of
e-Science, helping the scientist in modeling the more
suitable scientific workflow for the experiment to be
executed.
The OWL ontology myGrid (Wolstencroft et al.,
2007) was modeled for discovering and composition
of web services in Bioinformatics domain using the
Taverna SWfMS (Oinn et al., 2004) with semantic an-
notations, where you can use inference to find com-
mon ancestors to the activities of workflows. The
myGrid ontology models knowledge into scientific
workflows based on super-classes algorithm, date,
metadata, task, data resource, file formats and ser-
vice. One of the drawbacks of this approach is that
it does not present a clear separation between classes
related to modeling and visualization for the domain
addressed. SW-Ontology seeks to extend the domain
of scientific workflows, including a clear separation
between view and modeling classes, with the possi-
bility of expanding the scope of ontology for several
similar software systems.
In (Fox et al., 2009) is presented a semantic data
framework that models an OWL-DL ontology for the
representation of knowledge in the sub-domain of
Physics related to the Sun and the Earth, describing
concepts, relations and attributes of physical mag-
nitudes. The ontology is divided into main classes
Instrument, Observatory, Operating Mode, Parame-
ter, Coordinate and Data Archive. In (Oliveira et al.,
2009) is presented an ontology for the semantic mod-
eling of scientific workflows related to oil exploration
in deep waters. The ontology was used to define
some semantic concepts in order to provide support
for workflow composition. A case study is discussed
and, according to the authors, the results reinforce the
benefits of semantic support during the manual chain-
ing of processes and subworkflows. In both works,
the emphasis is on classes related to models. None of
them presents classes related to the implementation of
a scientific workflow.
2.4 Scope of the Work
SW-Ontology aims to describe the knowledge rep-
resented in scientific workflows, emphasizing such
modeling in the context of SWfMS Vistrails. In ad-
dition, SW-Ontology tries to incorporate to Vistrails
semantic modeling facilities such as resources for
queries and analysis of data provenance.
Vistrails represents a scientific workflow as an
acyclic graph. This SWfMS gives great emphasis on
data and process provenance and allows comparisons
of results to generate complex views. The choice
for this SWfMS was based on our group interest in
data provenance and Vistrails has an interesting data
provenance mechanism. Besides, Vistrails is related
with other developing works at the Research Center
on Software Quality (NPQS) of Federal University of
Juiz de Fora (UFJF) that uses this environment for sci-
entific workflows design and implementation. Cur-
rently, the NPQS focuses on the provision of an in-
frastructure for e-Science, named ASOW-Science
ICEIS 2010 - 12th International Conference on Enterprise Information Systems
116
(Matos et al., 2009) that includes technologies like on-
tologies, components and agents, with applications in
Bioinformatics, Agriculture and Education.
The SW-Ontology was developed using OWL-
DL, which ensures high capacity of expressiveness
and the inference computability in a finite time. The
adoption of OWL-DL is reinforced by the current sta-
tus of the language that is recommended by W3C
Consortium as part of a set of technologies for the
development of Semantic Web. The prototype im-
plementation was done in the Prot
´
eg
´
e, an environ-
ment for creating and editing ontologies and knowl-
edge bases (Horridge, 2009).
3 SEMANTIC MODELING OF A
SWfMS
For SW-Ontology development, we chose to use
a design pattern named Model-View (MV), which
is a derivation of the design pattern Model-View-
Controller (MVC). The purpose of the MVC pattern
arose from the increase growing of software devel-
opment complexity. In this context, it is essential
to separate the data (model), the layout (view) and
the control (controller). Thus, the implementation of
the MVC design pattern allows that changes in layout
does not affect the data and them the data can be reor-
ganized without significant changes in layout. As part
of this work, the MV design pattern is used in order to
separate the knowledge on the semantics of scientific
workflows design and implementation (model) from
the human-computer interaction performed by using
the SWfMS graphical user interface (view).
The use of MV is also important on the categoriza-
tion of classes, because it clearly defines the scope of
the model. It is still possible to glimpse the defini-
tion of different ontology view for a single ontology
model. Specifically in the context of this work, it is
possible to build view classes for the modeling do-
main from other SWfMS such as Taverna (Oinn et al.,
2004) and Kepler (Lud
¨
ascher et al., 2006), to name a
few.
In SW-Ontology, the subclasses model has been
divided in two hierarchies, i.e., as shown in Fig-
ure 1, the Model DomainEntity hierarchy for classes
of model and the ViewEntity for classes of view.
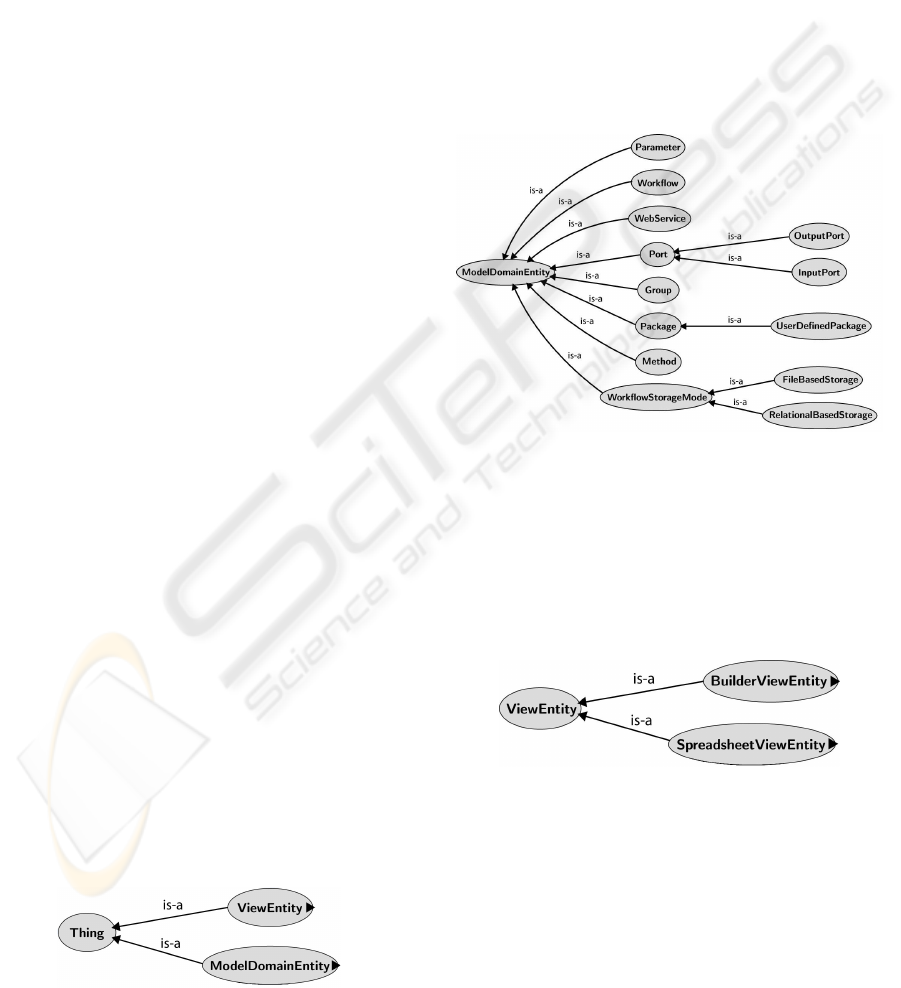
Figure 1: SW-Ontology main classes.
The ModelDomainEntity hierarchy is shown in
Figure 2. In Vistrails, a workflow can be defined as
a set of interconnected modules (class Module). The
links between modules are made from input and out-
put ports (class Port) and the actions taken by a mod-
ule are processed by methods (class Method) which
may receive parameters (class Parameter) for its im-
plementation. The various modules provided by Vis-
trails or by third parties are categorized into packages
(class Package). Vistrails also allows access to Web
services (class WebService) in the workflows compo-
sition. Finally, considering that this is an environment
for collaborative scientific exploration, it allows users
to store not only stand-alone files but also data from
relational databases (class Workflow StorageMode).
Figure 2: Model class hierarchy from ModelDomainEntity.
The view class ViewEntity describes the knowl-
edge of Vistrails users interaction environment (Fig-
ure 3). BuilderViewEntity represents the set of graph-
ical interfaces used for composition, execution and
query within scientific work-flows. SpreadsheetView
models the knowledge related to results visualization
and exploitation.
Figure 3: View class hierarchy from ViewEntity.
The current version of SW-Ontology contains 68
classes, 38 object properties (properties that indicate
a relationship between two classes) and 15 data prop-
erties (properties that indicate a relationship between
instances of classes and literals expressed in RDF or
XML Schema data types). All classes have annota-
tions comment type, whose goal is to provide a de-
scription of the knowledge modeled.
Figure 4 shows the representation of the model
SW-ONTOLOGY - A Proposal for Semantic Modeling of a Scientific Workflow Management System
117
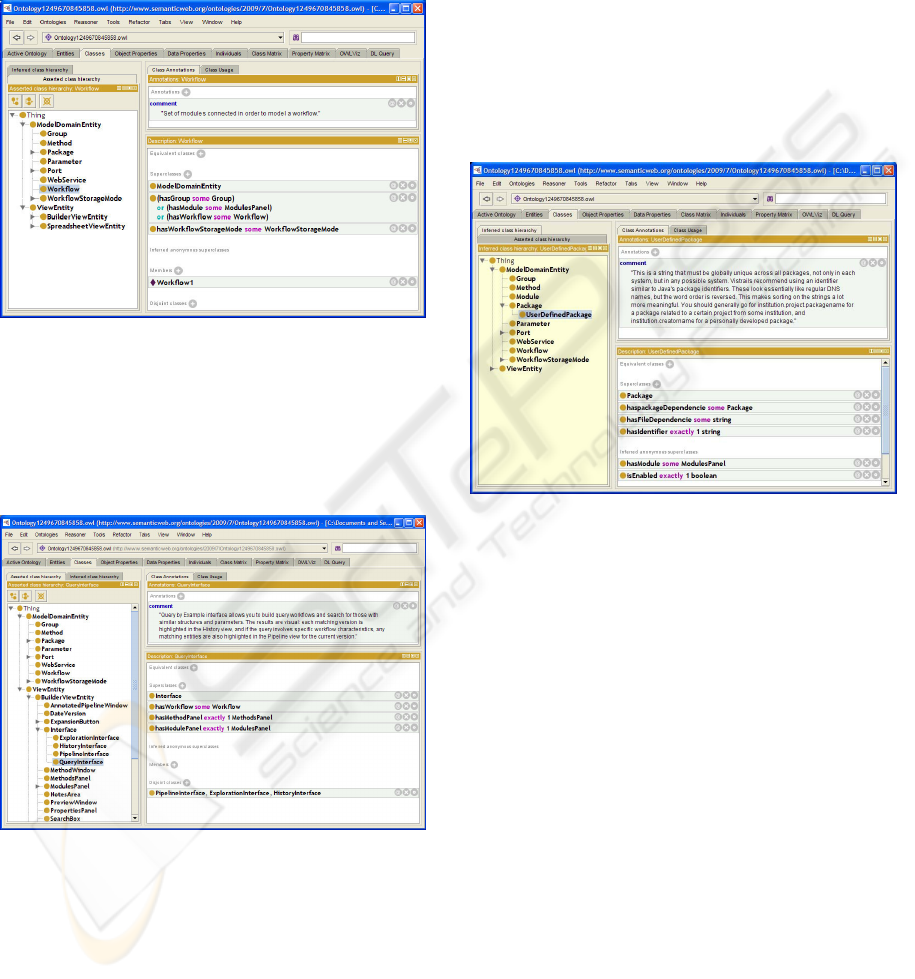
class Workflow on Prot
´
eg
´
e, defined as a set of inter-
connected modules in order to model a workflow. One
can observe that the adoption of the MV design pat-
tern allows that we can explicitly present the super
class relationship between Model DomainEntity and
Workflow class.
Figure 4: Modeling the model class Workflow on Prot
´
eg
´
e.
Figure 5 shows the view class QueryInterface,
which models the knowledge related to the definition
of queries such as query-by-example in a predefined
workflow that can locate and display graphically sub-
workflows or modules that meet the query performed.
Figure 5: Modeling the model class QueryInterface.
As an example of some restriction modeled in
SW-Ontology, we can see that the restriction has-
Workflow some Workflow relates an individual from
model class Workflow to the view class QueryInter-
face according to the property has-Workflow. This re-
striction may be interpreted as: it is necessary the ex-
istence of a previously constructed workflow in order
to run a query in the graphical interface. Thus, the
use of restrictions configures itself in a mechanism
capable of providing the connection between model
and view classes.
The Pellet (Sirin et al., 2007) was used for the
class hierarchy inference and SW-Ontology consis-
tency check. The use of an inference engine al-
lows you to extract new knowledge from the ontol-
ogy model built. Figure 6 illustrates results presented
for UserDefinedPackage, a ModelDomainEntity sub-
class, after SW-Ontology classification by Pellet. You
can verify that the reasoner has derived the restric-
tions HasModule some ModulesPanel and boolean
isEnabled exactly 1 from the class hierarchy built by
the classifier and their descriptions.
Figure 6: Modeling the model class UserDefinedPackage.
4 COMPOSITION OF
SCIENTIFIC WORKFLOWS:
USE OF SW-ONTOLOGY
ASOW-Science is a framework based on semantic
web services to compose workflows in a scenario of
e-Science. Can be understood as the specification and
development of an infrastructure whose purpose is to
provide computational support for researchers who
want to share experiments and results in a given ap-
plication domain (Silva et al., 2009). Specifically,
ASOW-Science manages the storage of ontologies
and semantic web services in distributed repositories,
and provide resources to the scientist to perform se-
mantic queries to the database. Furthermore, it is able
to make an automatic analysis of the services discov-
ered, in order to obtain possible compositions that can
be used to design workflows in a SWfMS.
Figure 7 represents the ASOW-Science layers.
The framework has two components: a client compo-
nent that invokes the service, and a middleware com-
ponent. The middleware consists of four layers:
ICEIS 2010 - 12th International Conference on Enterprise Information Systems
118
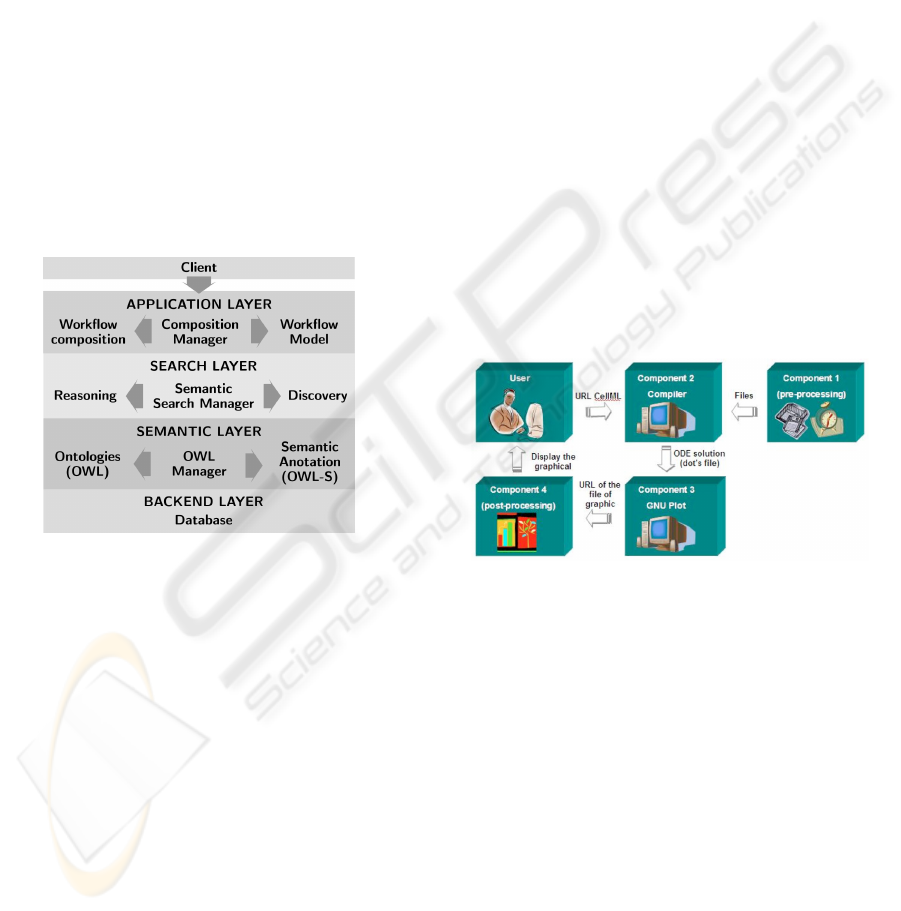
• the Backend Layer contains the database for stor-
age and query ontologies of the domain and se-
mantic annotations of Web services;
• the Semantic Layer is intended to manage the pro-
cesses of storage and query ontologies and seman-
tic annotations of services;
• the Search Layer performs the semantic search
and discovery of services according to the scien-
tist specifications. The information provided by
scientist and obtained by inferences is used to per-
form semantic search in the repositories to find
web services semantically compatible with each
task in the workflow. To find services, semantic
descriptions of services available in repositories
are analyzed and compared with the semantic data
related to each task;
• The Application Layer is responsible for model-
ing candidate compositions of scientific workflow
using the services discovered by the search layer.
Figure 7: The proposed framework architecture.
The integration of SW-Ontology with ASOW-
Science aims to provide scientists with a tool that fa-
cilitates the orchestration of a computer simulation in
the context of e-Science. After selecting the ontology
in the framework, the researcher can view the classes
and restrictions available, along with their semantic
descriptions. The ultimate goal of ASOW-Science is
to provide the scientists a search engine for semantic
Web services that exist in the framework repositories
and capable of performing the tasks selected from the
ontology.
To test ideas, we built a prototype that executes
a scientific workflow related to cell models specified
in CellML language (Matos et al., 2010). This work-
flow, related to the field of cardiac electrophysiology,
and developed at the UFJF Laboratory of Compu-
tational Physiology and High-Performance Comput-
ing, has several variations in terms of tools to add to
it. Thus, the proposed framework could be used to
semantically discover Web services which are more
suitable for the workflow implementation.
In this context, SW-Ontology and CELO (the do-
main ontology) are used together by the ASOW-
Science framework to provide the relationships
among the types of components selected by the re-
searcher and build a scientific workflow capable of
meeting the requirements.
In Figure 8 we have the execution schema of this
scientific workflow. Component 1 gets the system
date and current time, concatenate them in a sequence
of characters and create the files necessary to exe-
cute Component 2, using the sequence of characters
formed from the date of the system in their names.
Component 2 encapsulates a compiler that generates
an executable C code from a CellML model (Beard
et al., 2009), and executes the code to obtain output
data — the solution of an ordinary differential equa-
tion (ODE). Component 3 encapsulates a tool capa-
ble of generating a graph from a text file containing
ordered pairs. This component receives as input the
output file of Component 2 and creates a file to the
generated graph. Its output is the URL of the graph
file. Finally, Component 4 displays the graphical so-
lution of the ODE.
Figure 8: Cardiac electrophysiology workflow schema.
Considering the workflow shown in Figure 8,
classes of SW-Ontology as Workflow, Module and
ConcatenateStringModule, among others, represent
the related knowledge which constitutes the basis for
the composition of a scientific workflow within the
prototype.
Currently, there is a prototype of the proposed ar-
chitecture, developed as a Web service. This proto-
type allows that the scientist choose terms from SW-
Ontology and CELO ontology and links these terms
to tasks that must compose the workflow. Analyz-
ing the semantic annotation of Semantic Web services
that are stored in a repository, these terms could be
used to discover services that best fit the model pro-
vided by the user.
SW-ONTOLOGY - A Proposal for Semantic Modeling of a Scientific Workflow Management System
119

5 FINAL CONSIDERATIONS
Researches in e-Science have gained increase rele-
vance. However, there are few studies in the field
of Software Engineering focused on the topic and, in
particular, as in the use of ontologies in the e-Science
researches (Palazzi et al., 2009).
This paper proposes a model of semantic descrip-
tion based on OWL-DL ontologies to describe the
knowledge related to scientific workflow design and
implementation. Using the MV design pattern, an
ontology was built, separating the concepts related
to the semantic of scientific workflows orchestration
(model) and from the human-computer interaction
as of the computing environment graphical user in-
terface used (view). Currentlly, we are using SW-
Ontology in a huge project related to scientific work-
flow specification in the human diseases domain.
REFERENCES
Barga, R. S. and Digiampietri, L. A. (2008). Auto-
matic capture and efficient storage of e-science exper-
iment provenance. Concurr. Comput. : Pract. Exper.,
20(5):419–429.
Beard, D. A., Britten, R., Cooling, M. T., Garny, A., Hal-
stead, M. D. B., Hunter, P. J., Lawson, J., Lloyd,
C. M., Marsh, J., Miller, A., Nickerson, D. P., Nielsen,
P. M. F., Nomura, T., Subramanium, S., Wimalaratne,
S. M., and Yu, T. (2009). Cellml metadata standards,
associated tools and repositories. Physical and Engi-
neering Sciences, 367(1895):1845–1867.
Fox, P., McGuinness, D. L., Cinquini, L., West, P., Gar-
cia, J., Benedict, J. L., and Middleton, D. (2009).
Ontology-supported scientific data frameworks: The
virtual solar-terrestrial observatory experience. Com-
put. Geosci., 35(4):724–738.
Gruber, T. R. (1993). A translation approach to portable on-
tology specifications. Knowl. Acquis., 5(2):199–220.
Hewlett-Packard (2009). Jena semantic web framework.
Horridge, M. (2009). Prot
´
eg
´
e owl tutorial. Technical Report
v.1.2, The University of Manchester.
Lud
¨
ascher, B., Altintas, I., Berkley, C., Higgins, D., Jaeger,
E., Jones, M., Lee, E. A., Tao, J., and Zhao, Y. (2006).
Scientific workflow management and the kepler sys-
tem. Concurr. Comput. : Pract. Exper., 18(10):1039–
1065.
Matos, E. E., Mendes, L. F., Campos, F., and Braga, R.
(2009). Asow-science: a service oriented framework
to support e-science applications. In IRI’09: Proceed-
ings of the 10th IEEE international conference on In-
formation Reuse & Integration, pages 53–56, Piscat-
away, NJ, USA. IEEE Press.
Matos, E. E. E., Campos, F., Braga, R., and Palazzi, D.
(2010). Celows: an ontology based framework for
the provision of semantic web services related to bi-
ological models. Journal of biomedical informatics,
43(1):125–136.
Menager, H. and Lacroix, Z. (2006). A workflow engine for
the execution of scientific protocols. In ICDEW ’06:
Proceedings of the 22nd International Conference on
Data Engineering Workshops, page 68, Washington,
DC, USA. IEEE Computer Society.
Oinn, T., Addis, M., Ferris, J., Marvin, D., Carver, T.,
Pocock, M. R., and Wipat, A. (2004). Taverna: A tool
for the composition and enactment of bioinformatics
workflows. Bioinformatics, 20:3045–3054.
Oliveira, D. d., Cunha, L., Tomaz, L., Pereira, V., and Mat-
toso, M. (2009). Using ontologies to support deep wa-
ter oil exploration scientific workflows. In SERVICES
’09: Proceedings of the 2009 Congress on Services -
I, pages 364–367, Washington, DC, USA. IEEE Com-
puter Society.
Palazzi, D., Silva, L., Mendes, L., Gaspar, W., Matos, E.,
Campos, F., and Braga, R. (2009). Using ontologies
in e-science projects (in portuguese). In II Seminar on
Ontology Research in Brazil.
Silva, L., Campos, F., and Braga, R. (2009). A framework
for semantic composition of scientific workflows. In
IADIS’09: Proceedings of the International Confer-
ence WWW/Internet.
Sirin, E., Parsia, B., Grau, B. C., Kalyanpur, A., and Katz,
Y. (2007). Pellet: A practical owl-dl reasoner. Web
Semantics, 5(2):51–53.
Tsarkov, D. and Horrocks, I. (2006). Fact++ description
logic reasoner: System description. In Proc. of the Int.
Joint Conf. on Automated Reasoning (IJCAR 2006),
volume 4130 of Lecture Notes in Artificial Intelli-
gence, pages 292–297. Springer.
Wolstencroft, K., Alper, P., Hull, D., Wroe, C., Lord, P. W.,
Stevens, R. D., and Goble, C. A. (2007). The my-
grid ontology: bioinformatics service discovery. Int.
J. Bioinformatics Res. Appl., 3(3):303–325.
Wroe, C., Goble, C., Goderis, A., Lord, P., Miles, S., Pa-
pay, J., Alper, P., and Moreau, L. (2007). Recycling
workflows and services through discovery and reuse:
Research articles. Concurr. Comput. : Pract. Exper.,
19(2):181–194.
ICEIS 2010 - 12th International Conference on Enterprise Information Systems
120
