
DIFFUSE MATRIX
An Optimized Data Structure for the Storage and Processing
of Hyperspectral Images
Jose M. Chaves-González, Miguel A. Vega-Rodríguez, Pablo J. Martínez-Cobo
Juan A. Gómez-Pulido and Juan M. Sánchez-Pérez
Univ. Extremadura, Dept. Technologies of Computers and Communications
Escuela Politécnica, Campus Universitario s/n, 10071, Cáceres, Spain
Keywords: Hyperspectral data format, diffuse matrix, storage optimization, geo-rectification, AVIRIS images.
Abstract: This paper proposes a new format for storing and processing hyperspectral images captured by spectrometer
AVIRIS (Airborne Visible/InfraRed Imaging Spectrometer). Obtaining such images is difficult, because the
sensor that takes the images is carried in an aircraft that suffers turbulences while the camera is taking
photos. So, a geo-rectification process is necessary to correct the information of different bands. The format
proposed in this paper, DMF (Diffuse Matrix Format), allows a more efficient storage, because a list with
the original information received in the sensor is saved for each position (X,Y) of the scanned ground. The
format of the list saves space and time because no redundant information is saved using it. To show the
possibilities of this new format an application that makes some thresholding and filter operations has been
built. This program, firstly, creates the diffuse matrix in memory from the file that stores the image
information, and then, some filter operations are executed over the diffuse matrix to check it. In this way,
we prove that diffuse matrix processing is fast and simple, as well as the space used in the disk for its
storage is quite less than the space used by typical formats.
1 INTRODUCTION
Typical raster hyperspectral image formats used in
remote sensing (BSQ, BIL, BIP, HDF-EOS, Geo-
TIFF… (HDF-EOS, 2006)) save redundant
information when they store an image with some
acquisition errors (which is very common).
Figure 1: Diagram of hyperspectral image acquisition by
AVIRIS spectrometer.
For this reason, the files that hold such images take
up a lot of space in the hard disk, or at least, more
space than the space necessary. Moreover, the image
processing is not very fast and requires a lot of
memory, because the typical matrix that holds a
hyperspectral image has to be entirely read before
any processing over it.
Figure 2: AVIRIS hyperspectral image before (left) and
after (right) geo-rectification.
The over-information of typical hyperspectral
image formats is caused because the original images,
which are taken using spectrometer AVIRIS
(AVIRIS, 2007) (as it can be seen in figure 1), have
to be pre-processed before any usage with them
(Martínez et al., 2005)(Brunn et al., 2003). This is
39
M. Chaves-González J., A. Vega-Rodríguez M., J. Martínez-Cobo P., A. Gómez-Pulido J. and M. Sánchez-Pérez J. (2007).
DIFFUSE MATRIX - An Optimized Data Structure for the Storage and Processing of Hyperspectral Images.
In Proceedings of the Second International Conference on Signal Processing and Multimedia Applications, pages 39-44
DOI: 10.5220/0002130800390044
Copyright
c
SciTePress

due to the turbulences suffered by the plane or other
problems that happen while the sensor is taken the
images. Figure 2 shows a hyperspectral image taken
with AVIRIS before and after its geo-rectification.
As we can see in that figure, before rectification, the
image seems to be deformed, because some pixels of
some bands are not in their appropriate place. After
the geo-rectification, the imperfections in the image
are corrected (misplaced pixels are interpolated
using their neighbours).
If we choose an image format that applies geo-
rectification we will have some remarkable
disadvantages, because the process that corrects the
image interpolates the missing or misplaced pixels
with their neighbouring pixels, and this causes that
some information is duplicated, or even erroneous,
in the file that contains the image.
Therefore, the disadvantages of geo-rectification
in the traditional three-dimension formats (Lx,y,l)
carry us to study and develop other ways of
representation, storage and analysis of AVIRIS
hyperspectral images, where any interpolated
information is not saved (so, any redundant or
erroneous information will be not stored in the file).
Some studies to improve hyperspectral data
formats have been published before. It is worth
mentioning Boardman work (Boardman, 1999). He
describes three types of file (IMG, GLT and GEO).
These files are created in the image geo-rectification
process, and they split up the image information
(between data and meta-data information), but this
work is different to ours, because we do not apply
any geo-rectification for the image storage, but when
the image is going to be processed.
The rest of the paper is organized as follows:
section 2 explains the solution suggested in this
work (diffuse matrix), after that we put forward the
application created for this study which works with
the new structure and then, in section 4, we speak
about the obtained results. Finally, conclusions and
future work are expounded in section 5.
2 NEW DATA STRUCTURE:
DIFFUSE MATRIX
There are some criteria that make an image format to
be a good format (Folk, 1998). Between this
characteristics can be pointed out the following:
The space that it occupies, both in disk and in
memory.
The easiness of the format (it has to be simple
and easy to understand).
It has to be self-describing, using a metadata
file or similar.
Allow sequential access through the image.
Information access has to be easy to implement.
Rigorous and perfectly clear definition.
It has to be efficient: using the format, image
processing algorithms has to run efficiently
and quickly.
We try to follow all the previous requirements in
our work. Besides, we have made a robust format
taking into account the problems caused by the
sensor characteristics (Martínez et al., 2005) (which
are explained in the introduction). Diffuse matrix
separates physical storage and logical processing.
Thus, spectral information is compressed and saved
using a file named DMF (Diffuse Matrix File). In
this file no redundant information is saved, and only
when the structure is loaded in memory (when the
image is going to be processed, but not in its
storage), the required operations to put in order the
spectrum in the matrix are done.
To preserve the acquisition scheme, and to have
the information necessary for each band anytime,
each position in the diffuse matrix (once it is loaded
in memory) is a pointer to a dynamic list, where
each node contains the information shown in fig. 3.
Figure 3: Data register created for each measurement
taken by the sensor.
As it can be seen in figure 3, each register has 6
fields: A pair of fields of two bytes for latitude (Lat.)
and longitude (Lon.) of the scanned area; another
two fields for incidence angle (I.A.) and observation
angle (O.A.); one field of one byte which holds the
pixel spatial resolution (Foot_print size) for the
captured image, and finally, a field of variable
length for the spectrum (data) scanned by the sensor.
In diffuse matrix, each cell contains a pointer,
which points to null if no information is hold for that
coordinate in the matrix (as we said, this structure
saves space, because there are not interpolation of
missing information. If there are nothing, it is saved
nothing); or points to a dynamic list where each
element is a register like the described in fig. 3.
SIGMAP 2007 - International Conference on Signal Processing and Multimedia Applications
40

Figure 4: Diffuse matrix structure with some example
nodes (empty and with information nodes).
Figure 4 shows the diffuse matrix structure. At
first sight, the advantages obtained with the usage of
this structure are:
Diffuse matrix does not need any information
not captured by the sensor, so, any
interpolation or processing is not needed to
save the information.
Original coordinates (latitude and longitude)
are saved for each measurement.
Data distribution is independent of
visualization. So, when data are shown, they
have to be organized before, because the
information is saved in an optimal way,
without considering data spatial location.
(Only in the latest stages of data processing,
the interpolation of the image is necessary -to
calculate missing data in the image
acquisition-).
3 AN EXAMPLE OF
APPLICATION FOR DIFFUSE
MATRIX PROCESSING
We have developed a program just to show the
possibilities and the usage of diffuse matrix. The
application fulfils the following goals:
Diffuse matrix generation from DMF file which
contains it.
Image visualization using diffuse matrix
structure.
Performance of some image processing
algorithms over the diffuse matrix (we have
chosen a set of thresholding and filter
operations).
3.1 Diffuse Matrix Loading from DMF
File
A DMF image is divided into two files: a DMF file,
which contains the image, and a HDR file (header
file), which contains meta-information about the
image. Fig. 5 shows the information held in a typical
HDR file. HDR file contains information about the
number of image samples (which will be the number
of cells with information in the diffuse matrix), the
number of rows and columns that the matrix will
have, the number of bands per sample (bands in the
image), and some values to calculate the exact
position (X,Y) of each sample.
Figure 5: HDR file with meta-information about a DMF
image.
To load the diffuse matrix from a DMF file is
necessary to use the HRD file (fig. 5) which is
associated with that DMF file. Loading the image in
the matrix basically consists in reading each sample
sequentially from the DMF file (we know how many
samples there are from HRD file). After reading one
sample, its position is calculated and that sample is
added to the right coordinate in the matrix.
Interpolation will be performed to calculate the final
position of each sample in the matrix if the image is
loaded in memory with some association factor
among the cells. The application allows 4 levels of
association (the user chooses it when the image is
loaded), as we will explain in the following section.
Diffuse matrix structure is hold in memory all
the time while the application is working with the
image. Thus, the position of each sample is only
calculated once at the beginning of the process (not
for every image processing operation), doing it in an
efficient way.
When the process starts, the number of samples
for a particular cell of the matrix is unknown. Only
when the image is completely loaded, we know if in
a particular cell is held one, several or no samples.
So, a matrix of dynamic lists is the best structure to
hold the image in memory.
The position of a particular sample in the diffuse
matrix is obtained using the following equations:
posX=(Lat. - MinLatXPoint) * GIFOVLat
posY=(Lon - MinLonYPoint) * GIFOVLon
(1)
Lat (latitude) and Lon (longitude) values are read
for each sample (fig. 3), while the other values are
the same for all the samples in the image and are
read from HDR file (fig. 5).
DIFFUSE MATRIX - An Optimized Data Structure for the Storage and Processing of Hyperspectral Images
41

3.2 Image Visualization using Diffuse
Matrix Structure
When the hyperspectral image is shown, it is
considered as a set of n monochrome images (256
grey-levels each). The user chooses the band that
he/she wants to see and this band is shown as a grey
BMP image (in fact, the application allows saving
that image as a BMP image). The user also chooses
the association factor that the matrix cells will suffer
when the image is loaded at the beginning.
Association among cells is allowed because some
cells in the matrix are empty (without samples); so,
if the association is not done, some areas in the
image will not have any information (in this case the
image has some pixels without information, and the
application shows them in purple colour -they are
not processed-). Fig. 6 shows an image loaded with
and without cell association.
Figure 6: Image visualization with no association among
cells (a) and with a 2x2 association (b).
The application allows association factors of 1x1
(no association), 2x2 (groups of 4 cells), 3x3 (groups
of 9 cells) and 4x4 (groups of 16 cells). As we can
see in fig. 6, the more association factor that the
image has, the smaller it appears in the window
when it is shown (this reduction of dimensions has
some performance advantages, as we will see in the
next section). For the right location of each sample
in the matrix, the formulas shown in equation 1 are
divided by the association factor chosen by the user.
For example, the column occupied by a sample is
calculated using the updated formula: posX =
(Latitude - MinLatXPoint) * GIFOVLat /
association factor (the same that in equation 1 but
divided by the association factor).
With the association, it is difficult that any cell of
the matrix stays empty, because in case that the
association factor is, for instance, equal to 2, each
cell contains the samples of 4 neighbouring pixels.
3.3 Image Processing using Diffuse
Matrix
To prove the easiness and potential of diffuse
matrix, we have developed some thresholding and
filter operations (González and Woods, 1992). The
user uses the window shown in figure 7 to configure
the different operations that can be done over the
diffuse matrix.
In this window, the user can configure the band
which he/she wants to work with (on the right area
of the window) and the operation that he/she wants
to apply over the selected band (no operation -only
showing the image-, a thresholding operation or a
filter operation –of smoothing, sharpening or edge
detection-). In particular, we have implemented 15
filters: Average3x3, Average5x5, Average7x7,
Gauss0391, Gauss0625, AverageModA,
AverageModB, Laplace1, Laplace2, SobelX,
SobelY, Sobel45+, Sobel45-, Prewitt1 and Prewitt2.
In summary, we have implemented an important
amount of operations using the structure (both
thresholding operations and convolution operations)
to perform a complete test of the behaviour of the
diffuse matrix for these operations, which are quite
important in image processing. In fig. 8, some
examples of filtering are shown.
Figure 7: Image processing configuration window.
Moreover, if the user chooses association among
the cells of the matrix, an important advantage is
that the image which the application has to operate is
“a dense image”, smaller than the original image, so,
the image processing is faster and more efficient. In
fact, this idea is similar to the one used in Vector
Architectures, with the technique called “scatter-
gather” –which is included on many of the recent
supercomputers–, where data are grouped to process
them faster in a parallel way). In conclusion, diffuse
matrix can be processed faster than traditional
formats if the user chose association among the cells
when the image was loaded.
SIGMAP 2007 - International Conference on Signal Processing and Multimedia Applications
42
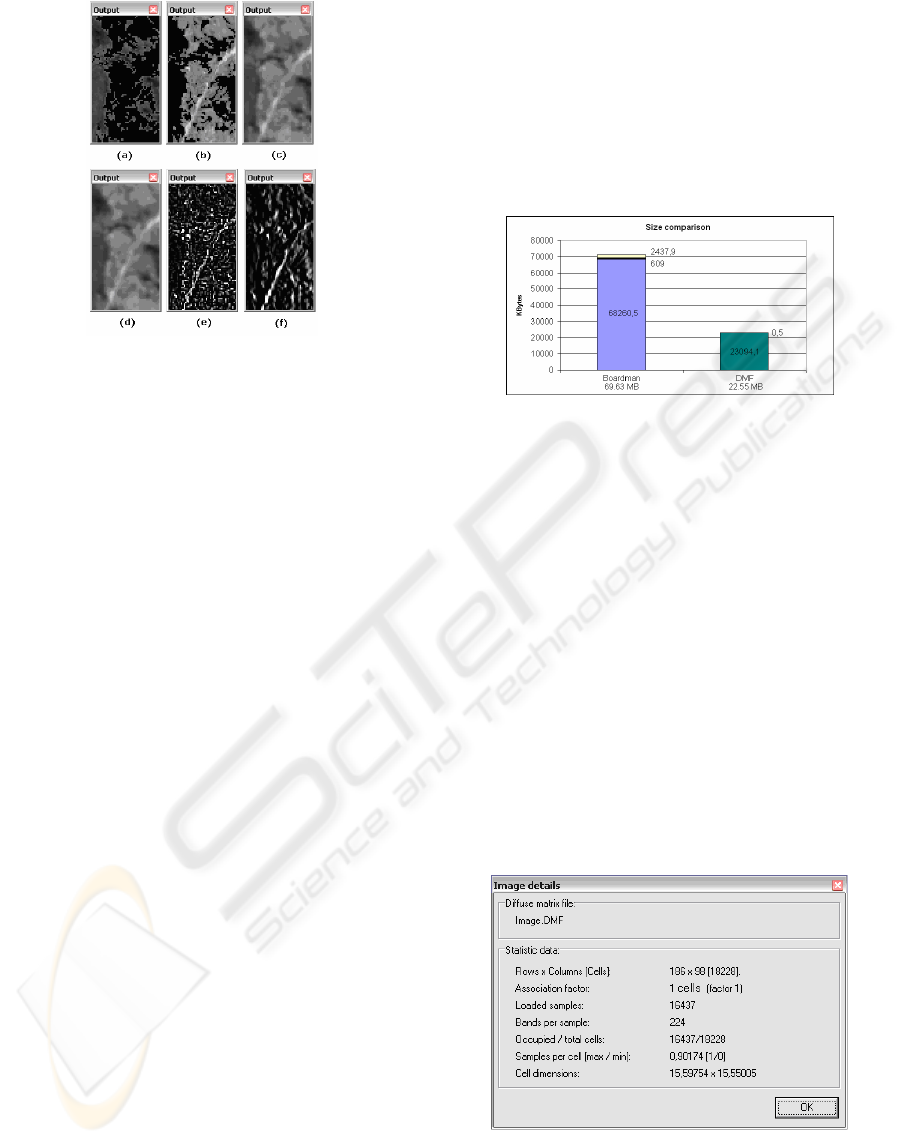
Figure 8: Some example of filters applied over the band 36
of the image.
The filters observed in figure 8 are: (a)
thresholding -pixels with values less than 100
become black-, (b) thresholding -pixels with values
greater than 100 become black-, (c) 3x3 average
filter, (d) Gauss filter, (e) Laplace filter, (f) SobelX
gradient filter.
We have used the following method to
implement every filter operations over the diffuse
matrix: For each cell in the matrix is calculated the
centre position and the distance from this centre to
every sample in the cell. Using these distances, the
weight of each sample over the pixel which is going
to be drawn in that position is calculated. The value
of the pixel associated to a cell is obtained taking
into account all the weights of all the samples in the
cell after a normalization process into the range 0-
255. This process is done for every cell in the
selected band. Finally, the chosen operation is
applied over the normalized pixel matrix and the
result is shown as can be seen in fig. 8.
4 SOME RESULTS OBTAINED
WITH THE STUDY
We have done some size comparisons, and we can
conclude that due to the lack of redundant
information included in the DMF file from geo-
rectification, the size of the image is much smaller
than using traditional formats. In fact, we have done
a comparison with the format proposed by
Boardman in (Boardman, 1999) (which is an
optimization that consist in 3 files to support an
image -GEO, GLT and UTM files-). Using the same
image (IVAHOHA_BEACH_low_altitude), the
following results are obtained: our file with the
diffuse matrix (DMF) occupies 23,648,372 bytes
(22.5 MB) and our HDR file takes up an almost
negligible 551 bytes. On the other hand, the
Boardman solution files take up: 69,898,752 bytes
(66.6 MB) for the GEO file, 624,096 bytes (609 KB)
for the GLT file and 2,496,384 bytes (2.4 MB) for
the UTM file. If we compare the results, the
compression ratio obtained is over 3:1, as we can see
in fig. 9.
Figure 9: Size comparison between Boardman format and
DMF format.
Furthermore, as we said in section 3.2, we can
select an association factor when the image is
loaded. If we associate cells when the matrix is
loaded, the matrix will be compacted in memory (it
has less dimensions) and the final size will be
reduced. In fig. 10 we show the details for the same
image without association factor and in figure 11
with association factor equal to 2. The diffuse matrix
dimensions with an association factor of 2 are
reduced to a quarter, but the cell dimensions are
bigger with the association, because there are more
points for each cell. Moreover, it is important to
point that, as it can be seen in figures 10 and 11,
with no association, the matrix has 0.9 samples per
cell (there was cells with no samples), and with the
association, there are 3.6 samples for each cell (at
least 1 sample per cell and no more than 4 samples).
Figure 10: Image details for an image loaded without any
cell association.
DIFFUSE MATRIX - An Optimized Data Structure for the Storage and Processing of Hyperspectral Images
43

Figure 11: Image details for an image loaded with cell
association (4 cells).
Anyway, with any association factor, the image
is processed in the same way, because the structure
of the matrix is always the same: a matrix of lists,
where each node in the list contains the information
of a sample acquired by the sensor, with altitude and
longitude data (fig. 3), so the processing of this
structure is homogeneous and efficient.
5 CONCLUSIONS AND FUTURE
WORK
We have described a new format to work with
AVIRIS hyperspectral images. With the DMF
format, the physical storage in the disk (DMF and
HDR files) and the later image processing (diffuse
matrix with or without association of cells) have
been clearly separated. This means that the same
images take much less space in hard disk using DMF
format than other typical formats. Furthermore, the
image can be processed in a more efficient way,
because it is loaded as a matrix of dynamic lists,
taking advantage of the structure used. We have
mentioned some of these advantages in the previous
sections, but moreover, the diffuse matrix makes
easier the parallelization of algorithms, because
when the image is going to be processed, each cell is
independent of the others in the matrix, and it is
possible to divide the diffuse matrix and process
each piece in a separate/parallel way, improving and
speeding up the image processing (this aspect is very
interesting because the size of hyperspectral images
is quite big).
In conclusion, this work is a first step (the most
important milestone) for the upcoming works in next
months. Future work includes: (a) a detailed
statistical comparison among different algorithms
and different formats to get the concrete
improvement obtained by the diffuse matrix in each
case; (b) a study in depth about the parallelization of
some algorithms using the proposed structure
(diffuse matrix).
ACKNOWLEDGEMENTS
This work has been developed in part thanks to the
OPLINK project (TIN2005-08818-C04-03).
REFERENCES
AVIRIS, Jet Propulsion Laboratory, NASA. AVIRIS,
http://aviris.jpl.nasa.gov/, 2007.
Boardman, J.W.. Precision Geocoding of Low-Altitude
AVIRIS Data: Lessons Learned in 1998. 8
th
Annual
JPL Airborne Geoscience Workshop, Pasadena,
California, USA, 1999.
Brunn, A., Fischer, C., Dittmann, C., Richter, R.. Quality
Assessment, Atmospheric and Geometric Correction of
Airborne Hyperspectral HyMap Data. 3
rd
EARSeL
Workshop on Imaging Spectroscopy, Herrsching,
Germany, May 2003.
Folk, M.. HDF as an Archive Format: Issues and
Recommendations. White paper, NCSA/University of
Illinois, http://hdf.ncsa.uiuc.edu/archive/
hdfasarchivefmt.htm, 1998.
González, R.C., Woods, R.E.. Digital Image Processing.
2
nd
Edition, Addison-Wesley, 1992.
HDF-EOS, NASA. HDF-EOS,
http://hdfeos.gsfc.nasa.gov/hdfeos/index.cfm, 2006.
Martínez, P.J., Hermosel, D., Green, R.O., Plaza, J., Pérez,
R.M.. An Improved Data Structure for AVIRIS-Type
Imaging Spectrometer Measurements. 13
th
JPL
Airborne Earth Science Workshop, Pasadena,
California, USA, May 2005.
SIGMAP 2007 - International Conference on Signal Processing and Multimedia Applications
44
